Akshay Deepak
λ-Scaled-Attention: A Novel Fast Attention Mechanism for Efficient Modeling of Protein Sequences
Jan 09, 2022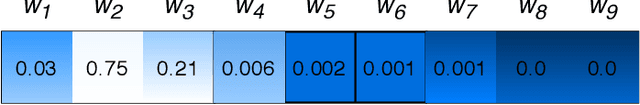
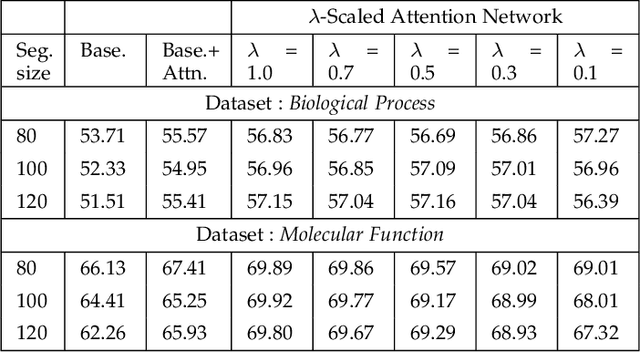
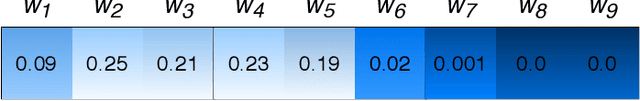
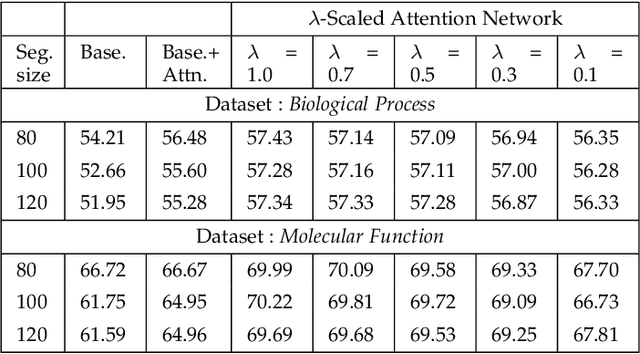
Abstract:Attention-based deep networks have been successfully applied on textual data in the field of NLP. However, their application on protein sequences poses additional challenges due to the weak semantics of the protein words, unlike the plain text words. These unexplored challenges faced by the standard attention technique include (i) vanishing attention score problem and (ii) high variations in the attention distribution. In this regard, we introduce a novel {\lambda}-scaled attention technique for fast and efficient modeling of the protein sequences that addresses both the above problems. This is used to develop the {\lambda}-scaled attention network and is evaluated for the task of protein function prediction implemented at the protein sub-sequence level. Experiments on the datasets for biological process (BP) and molecular function (MF) showed significant improvements in the F1 score values for the proposed {\lambda}-scaled attention technique over its counterpart approach based on the standard attention technique (+2.01% for BP and +4.67% for MF) and state-of-the-art ProtVecGen-Plus approach (+2.61% for BP and +4.20% for MF). Further, fast convergence (converging in half the number of epochs) and efficient learning (in terms of very low difference between the training and validation losses) were also observed during the training process.
Pitch-Synchronous Single Frequency Filtering Spectrogram for Speech Emotion Recognition
Aug 07, 2019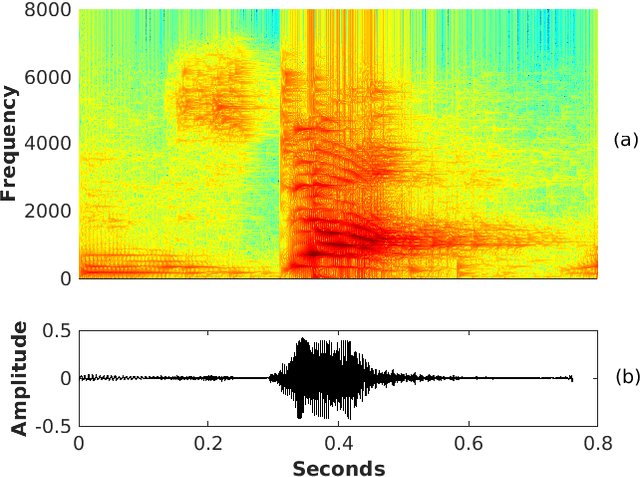
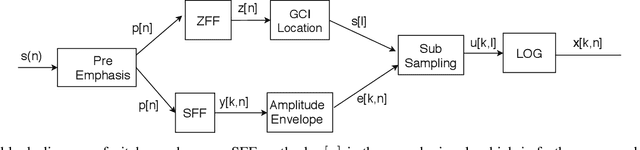
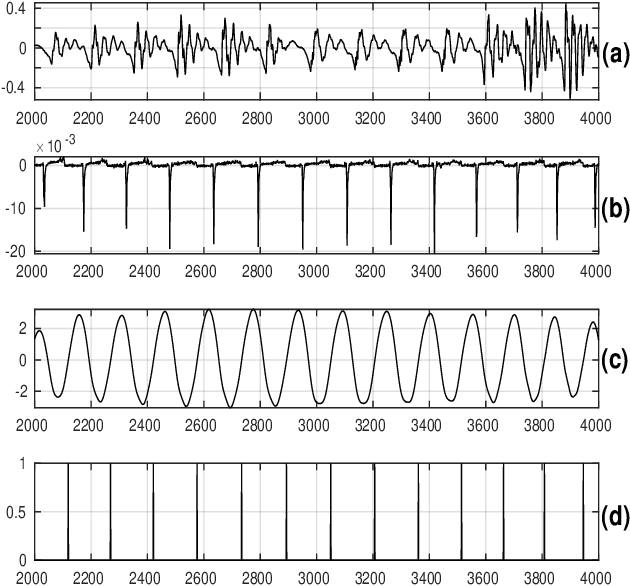
Abstract:Convolutional neural networks (CNN) are widely used for speech emotion recognition (SER). In such cases, the short time fourier transform (STFT) spectrogram is the most popular choice for representing speech, which is fed as input to the CNN. However, the uncertainty principles of the short-time Fourier transform prevent it from capturing time and frequency resolutions simultaneously. On the other hand, the recently proposed single frequency filtering (SFF) spectrogram promises to be a better alternative because it captures both time and frequency resolutions simultaneously. In this work, we explore the SFF spectrogram as an alternative representation of speech for SER. We have modified the SFF spectrogram by taking the average of the amplitudes of all the samples between two successive glottal closure instants (GCI) locations. The duration between two successive GCI locations gives the pitch, motivating us to name the modified SFF spectrogram as pitch-synchronous SFF spectrogram. The GCI locations were detected using zero frequency filtering approach. The proposed pitch-synchronous SFF spectrogram produced accuracy values of 63.95% (unweighted) and 70.4% (weighted) on the IEMOCAP dataset. These correspond to an improvement of +7.35% (unweighted) and +4.3% (weighted) over state-of-the-art result on the STFT sepctrogram using CNN. Specially, the proposed method recognized 22.7% of the happy emotion samples correctly, whereas this number was 0% for state-of-the-art results. These results also promise a much wider use of the proposed pitch-synchronous SFF spectrogram for other speech-based applications.
Deep Robust Framework for Protein Function Prediction using Variable-Length Protein Sequences
Nov 04, 2018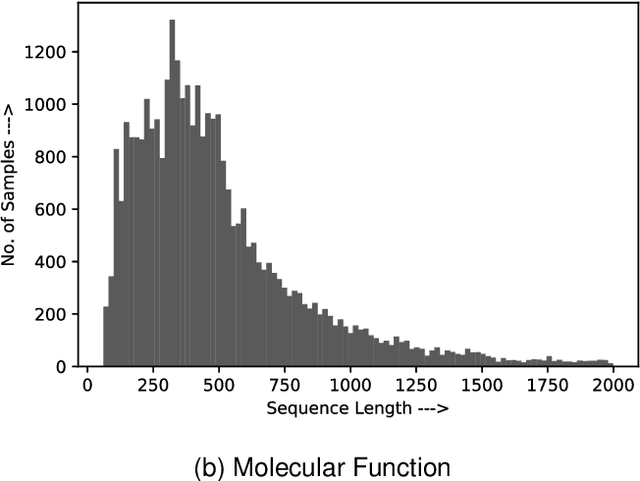
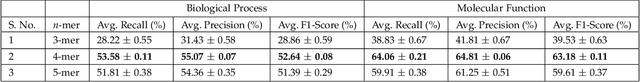
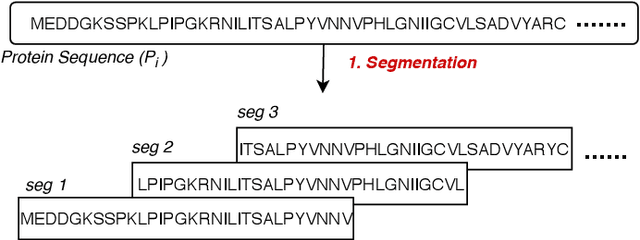
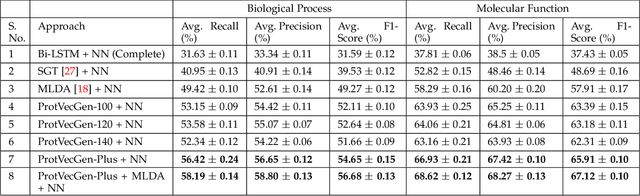
Abstract:Amino acid sequence portrays most intrinsic form of a protein and expresses primary structure of protein. The order of amino acids in a sequence enables a protein to acquire a particular stable conformation that is responsible for the functions of the protein. This relationship between a sequence and its function motivates the need to analyse the sequences for predicting protein functions. Early generation computational methods using BLAST, FASTA, etc. perform function transfer based on sequence similarity with existing databases and are computationally slow. Although machine learning based approaches are fast, they fail to perform well for long protein sequences (i.e., protein sequences with more than 300 amino acid residues). In this paper, we introduce a novel method for construction of two separate feature sets for protein sequences based on analysis of 1) single fixed-sized segments and 2) multi-sized segments, using bi-directional long short-term memory network. Further, model based on proposed feature set is combined with the state of the art Multi-lable Linear Discriminant Analysis (MLDA) features based model to improve the accuracy. Extensive evaluations using separate datasets for biological processes and molecular functions demonstrate promising results for both single-sized and multi-sized segments based feature sets. While former showed an improvement of +3.37% and +5.48%, the latter produces an improvement of +5.38% and +8.00% respectively for two datasets over the state of the art MLDA based classifier. After combining two models, there is a significant improvement of +7.41% and +9.21% respectively for two datasets compared to MLDA based classifier. Specifically, the proposed approach performed well for the long protein sequences and superior overall performance.
DNN-HMM based Speaker Adaptive Emotion Recognition using Proposed Epoch and MFCC Features
Jun 04, 2018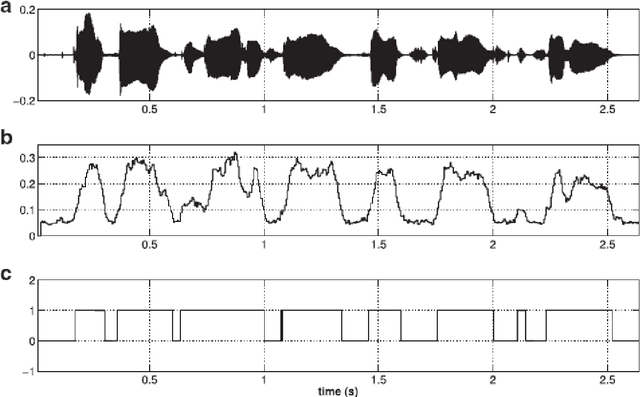
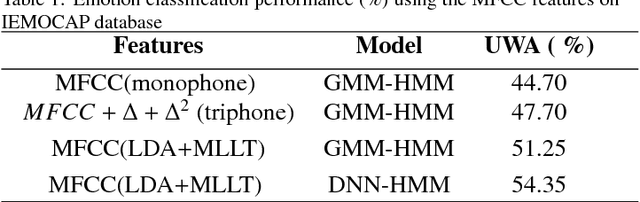
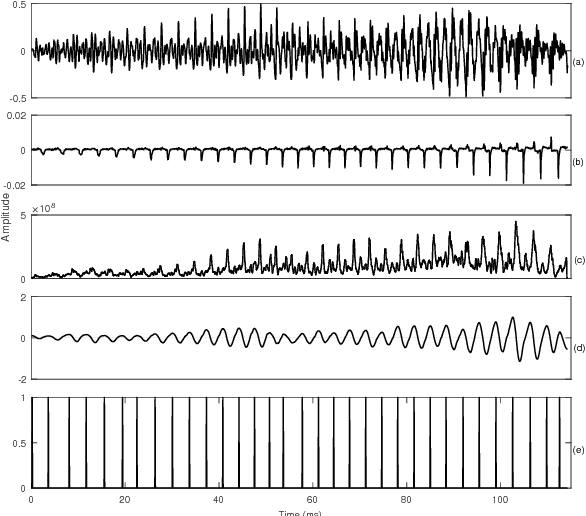
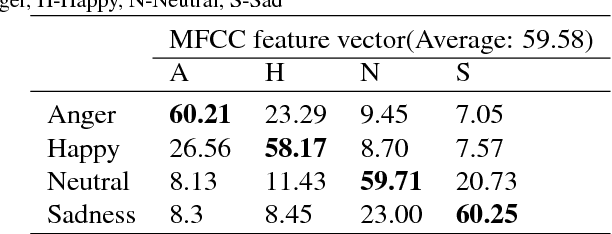
Abstract:Speech is produced when time varying vocal tract system is excited with time varying excitation source. Therefore, the information present in a speech such as message, emotion, language, speaker is due to the combined effect of both excitation source and vocal tract system. However, there is very less utilization of excitation source features to recognize emotion. In our earlier work, we have proposed a novel method to extract glottal closure instants (GCIs) known as epochs. In this paper, we have explored epoch features namely instantaneous pitch, phase and strength of epochs for discriminating emotions. We have combined the excitation source features and the well known Male-frequency cepstral coefficient (MFCC) features to develop an emotion recognition system with improved performance. DNN-HMM speaker adaptive models have been developed using MFCC, epoch and combined features. IEMOCAP emotional database has been used to evaluate the models. The average accuracy for emotion recognition system when using MFCC and epoch features separately is 59.25% and 54.52% respectively. The recognition performance improves to 64.2% when MFCC and epoch features are combined.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge