Anna Bernasconi
Capturing research literature attitude towards Sustainable Development Goals: an LLM-based topic modeling approach
Nov 05, 2024



Abstract:The world is facing a multitude of challenges that hinder the development of human civilization and the well-being of humanity on the planet. The Sustainable Development Goals (SDGs) were formulated by the United Nations in 2015 to address these global challenges by 2030. Natural language processing techniques can help uncover discussions on SDGs within research literature. We propose a completely automated pipeline to 1) fetch content from the Scopus database and prepare datasets dedicated to five groups of SDGs; 2) perform topic modeling, a statistical technique used to identify topics in large collections of textual data; and 3) enable topic exploration through keywords-based search and topic frequency time series extraction. For topic modeling, we leverage the stack of BERTopic scaled up to be applied on large corpora of textual documents (we find hundreds of topics on hundreds of thousands of documents), introducing i) a novel LLM-based embeddings computation for representing scientific abstracts in the continuous space and ii) a hyperparameter optimizer to efficiently find the best configuration for any new big datasets. We additionally produce the visualization of results on interactive dashboards reporting topics' temporal evolution. Results are made inspectable and explorable, contributing to the interpretability of the topic modeling process. Our proposed LLM-based topic modeling pipeline for big-text datasets allows users to capture insights on the evolution of the attitude toward SDGs within scientific abstracts in the 2006-2023 time span. All the results are reproducible by using our system; the workflow can be generalized to be applied at any point in time to any big corpus of textual documents.
Searching COVID-19 clinical research using graphical abstracts
Oct 06, 2023



Abstract:Objective. Graphical abstracts are small graphs of concepts that visually summarize the main findings of scientific articles. While graphical abstracts are customarily used in scientific publications to anticipate and summarize their main results, we propose them as a means for expressing graph searches over existing literature. Materials and methods. We consider the COVID-19 Open Research Dataset (CORD-19), a corpus of more than one million abstracts; each of them is described as a graph of co-occurring ontological terms, selected from the Unified Medical Language System (UMLS) and the Ontology of Coronavirus Infectious Disease (CIDO). Graphical abstracts are also expressed as graphs of ontological terms, possibly augmented by utility terms describing their interactions (e.g., "associated with", "increases", "induces"). We build a co-occurrence network of concepts mentioned in the corpus; we then identify the best matches of graphical abstracts on the network. We exploit graph database technology and shortest-path queries. Results. We build a large co-occurrence network, consisting of 128,249 entities and 47,198,965 relationships. A well-designed interface allows users to explore the network by formulating or adapting queries in the form of an abstract; it produces a bibliography of publications, globally ranked; each publication is further associated with the specific parts of the abstract that it explains, thereby allowing the user to understand each aspect of the matching. Discussion and Conclusion. Our approach supports the process of scientific hypothesis formulation and evidence search; it can be reapplied to any scientific domain, although our mastering of UMLS makes it most suited to clinical domains.
Exploring the evolution of research topics during the COVID-19 pandemic
Oct 05, 2023
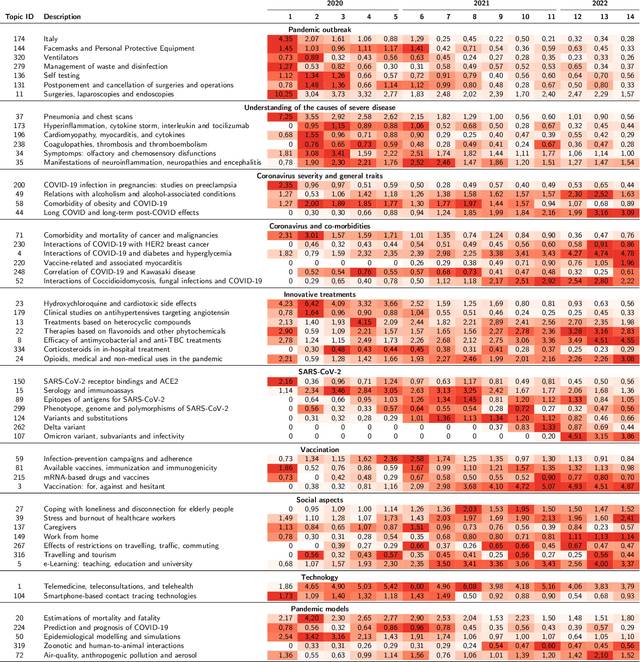


Abstract:The COVID-19 pandemic has changed the research agendas of most scientific communities, resulting in an overwhelming production of research articles in a variety of domains, including medicine, virology, epidemiology, economy, psychology, and so on. Several open-access corpora and literature hubs were established; among them, the COVID-19 Open Research Dataset (CORD-19) has systematically gathered scientific contributions for 2.5 years, by collecting and indexing over one million articles. Here, we present the CORD-19 Topic Visualizer (CORToViz), a method and associated visualization tool for inspecting the CORD-19 textual corpus of scientific abstracts. Our method is based upon a careful selection of up-to-date technologies (including large language models), resulting in an architecture for clustering articles along orthogonal dimensions and extraction techniques for temporal topic mining. Topic inspection is supported by an interactive dashboard, providing fast, one-click visualization of topic contents as word clouds and topic trends as time series, equipped with easy-to-drive statistical testing for analyzing the significance of topic emergence along arbitrarily selected time windows. The processes of data preparation and results visualization are completely general and virtually applicable to any corpus of textual documents - thus suited for effective adaptation to other contexts.
Quantum Clustering with k-Means: a Hybrid Approach
Dec 15, 2022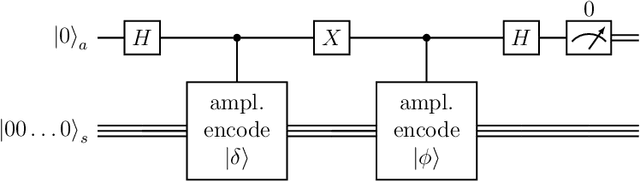
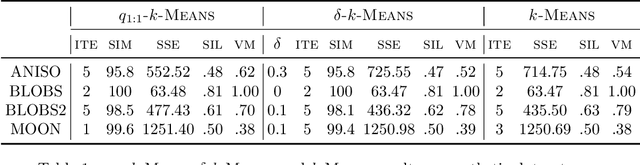
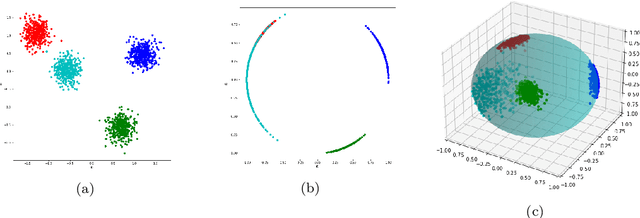
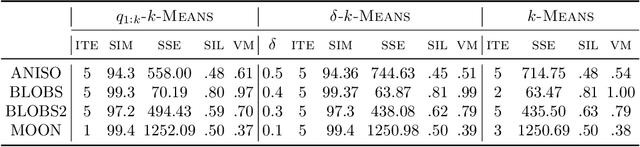
Abstract:Quantum computing is a promising paradigm based on quantum theory for performing fast computations. Quantum algorithms are expected to surpass their classical counterparts in terms of computational complexity for certain tasks, including machine learning. In this paper, we design, implement, and evaluate three hybrid quantum k-Means algorithms, exploiting different degree of parallelism. Indeed, each algorithm incrementally leverages quantum parallelism to reduce the complexity of the cluster assignment step up to a constant cost. In particular, we exploit quantum phenomena to speed up the computation of distances. The core idea is that the computation of distances between records and centroids can be executed simultaneously, thus saving time, especially for big datasets. We show that our hybrid quantum k-Means algorithms can be more efficient than the classical version, still obtaining comparable clustering results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge