Andrea Zanoni
On the performance of multi-fidelity and reduced-dimensional neural emulators for inference of physiologic boundary conditions
Jun 13, 2025Abstract:Solving inverse problems in cardiovascular modeling is particularly challenging due to the high computational cost of running high-fidelity simulations. In this work, we focus on Bayesian parameter estimation and explore different methods to reduce the computational cost of sampling from the posterior distribution by leveraging low-fidelity approximations. A common approach is to construct a surrogate model for the high-fidelity simulation itself. Another is to build a surrogate for the discrepancy between high- and low-fidelity models. This discrepancy, which is often easier to approximate, is modeled with either a fully connected neural network or a nonlinear dimensionality reduction technique that enables surrogate construction in a lower-dimensional space. A third possible approach is to treat the discrepancy between the high-fidelity and surrogate models as random noise and estimate its distribution using normalizing flows. This allows us to incorporate the approximation error into the Bayesian inverse problem by modifying the likelihood function. We validate five different methods which are variations of the above on analytical test cases by comparing them to posterior distributions derived solely from high-fidelity models, assessing both accuracy and computational cost. Finally, we demonstrate our approaches on two cardiovascular examples of increasing complexity: a lumped-parameter Windkessel model and a patient-specific three-dimensional anatomy.
Enabling stratified sampling in high dimensions via nonlinear dimensionality reduction
Jun 10, 2025Abstract:We consider the problem of propagating the uncertainty from a possibly large number of random inputs through a computationally expensive model. Stratified sampling is a well-known variance reduction strategy, but its application, thus far, has focused on models with a limited number of inputs due to the challenges of creating uniform partitions in high dimensions. To overcome these challenges, we perform stratification with respect to the uniform distribution defined over the unit interval, and then derive the corresponding strata in the original space using nonlinear dimensionality reduction. We show that our approach is effective in high dimensions and can be used to further reduce the variance of multifidelity Monte Carlo estimators.
NeurAM: nonlinear dimensionality reduction for uncertainty quantification through neural active manifolds
Aug 07, 2024Abstract:We present a new approach for nonlinear dimensionality reduction, specifically designed for computationally expensive mathematical models. We leverage autoencoders to discover a one-dimensional neural active manifold (NeurAM) capturing the model output variability, plus a simultaneously learnt surrogate model with inputs on this manifold. The proposed dimensionality reduction framework can then be applied to perform outer loop many-query tasks, like sensitivity analysis and uncertainty propagation. In particular, we prove, both theoretically under idealized conditions, and numerically in challenging test cases, how NeurAM can be used to obtain multifidelity sampling estimators with reduced variance by sampling the models on the discovered low-dimensional and shared manifold among models. Several numerical examples illustrate the main features of the proposed dimensionality reduction strategy, and highlight its advantages with respect to existing approaches in the literature.
A Probabilistic Neural Twin for Treatment Planning in Peripheral Pulmonary Artery Stenosis
Dec 01, 2023Abstract:The substantial computational cost of high-fidelity models in numerical hemodynamics has, so far, relegated their use mainly to offline treatment planning. New breakthroughs in data-driven architectures and optimization techniques for fast surrogate modeling provide an exciting opportunity to overcome these limitations, enabling the use of such technology for time-critical decisions. We discuss an application to the repair of multiple stenosis in peripheral pulmonary artery disease through either transcatheter pulmonary artery rehabilitation or surgery, where it is of interest to achieve desired pressures and flows at specific locations in the pulmonary artery tree, while minimizing the risk for the patient. Since different degrees of success can be achieved in practice during treatment, we formulate the problem in probability, and solve it through a sample-based approach. We propose a new offline-online pipeline for probabilsitic real-time treatment planning which combines offline assimilation of boundary conditions, model reduction, and training dataset generation with online estimation of marginal probabilities, possibly conditioned on the degree of augmentation observed in already repaired lesions. Moreover, we propose a new approach for the parametrization of arbitrarily shaped vascular repairs through iterative corrections of a zero-dimensional approximant. We demonstrate this pipeline for a diseased model of the pulmonary artery tree available through the Vascular Model Repository.
Minimax Classification with 0-1 Loss and Performance Guarantees
Oct 15, 2020

Abstract:Supervised classification techniques use training samples to find classification rules with small expected 0-1 loss. Conventional methods achieve efficient learning and out-of-sample generalization by minimizing surrogate losses over specific families of rules. This paper presents minimax risk classifiers (MRCs) that do not rely on a choice of surrogate loss and family of rules. MRCs achieve efficient learning and out-of-sample generalization by minimizing worst-case expected 0-1 loss w.r.t. uncertainty sets that are defined by linear constraints and include the true underlying distribution. In addition, MRCs' learning stage provides performance guarantees as lower and upper tight bounds for expected 0-1 loss. We also present MRCs' finite-sample generalization bounds in terms of training size and smallest minimax risk, and show their competitive classification performance w.r.t. state-of-the-art techniques using benchmark datasets.
Supervised classification via minimax probabilistic transformations
Apr 16, 2019
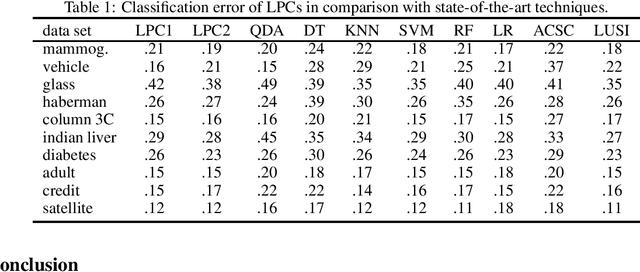
Abstract:Current leaning techniques for supervised classification consider classification rules in specific families and empirically quantify performance using test data. Selection among families of classification rules, techniques' choices, parameters, etc. is mostly guided by an experimentation stage in which the performance of different options is estimated by computing the corresponding empirical risks over a set of test examples. This empirically-based system design is inefficient and not robust since it requires to test a possibly large pool of choices and it relies in the reliability of the empirical performance quantification. This paper presents classification algorithms that we call linear probabilistic machines (LPMs) that consider unconstrained classification rules and obtain tight performance guaranties during learning. LPMs utilize polyhedral uncertainty sets that contain the actual probability distribution with a tunable confidence and are defined by a functional that assigns features-label pairs to real numbers. LPMs achieve smaller worst-case risk against such uncertainty sets than any classification rule and their performance can be efficiently upper and lower bounded without testing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge