Anders Blilie
Department of Pathology, Stavanger University Hospital, Stavanger, Norway, Faculty of Health Sciences, University of Stavanger, Stavanger, Norway
Validation of Diagnostic Artificial Intelligence Models for Prostate Pathology in a Middle Eastern Cohort
Dec 19, 2025Abstract:Background: Artificial intelligence (AI) is improving the efficiency and accuracy of cancer diagnostics. The performance of pathology AI systems has been almost exclusively evaluated on European and US cohorts from large centers. For global AI adoption in pathology, validation studies on currently under-represented populations - where the potential gains from AI support may also be greatest - are needed. We present the first study with an external validation cohort from the Middle East, focusing on AI-based diagnosis and Gleason grading of prostate cancer. Methods: We collected and digitised 339 prostate biopsy specimens from the Kurdistan region, Iraq, representing a consecutive series of 185 patients spanning the period 2013-2024. We evaluated a task-specific end-to-end AI model and two foundation models in terms of their concordance with pathologists and consistency across samples digitised on three scanner models (Hamamatsu, Leica, and Grundium). Findings: Grading concordance between AI and pathologists was similar to pathologist-pathologist concordance with Cohen's quadratically weighted kappa 0.801 vs. 0.799 (p=0.9824). Cross-scanner concordance was high (quadratically weighted kappa > 0.90) for all AI models and scanner pairs, including low-cost compact scanner. Interpretation: AI models demonstrated pathologist-level performance in prostate histopathology assessment. Compact scanners can provide a route for validation studies in non-digitalised settings and enable cost-effective adoption of AI in laboratories with limited sample volumes. This first openly available digital pathology dataset from the Middle East supports further research into globally equitable AI pathology. Funding: SciLifeLab and Wallenberg Data Driven Life Science Program, Instrumentarium Science Foundation, Karolinska Institutet Research Foundation.
Artificial Intelligence-Assisted Prostate Cancer Diagnosis for Reduced Use of Immunohistochemistry
Mar 31, 2025Abstract:Prostate cancer diagnosis heavily relies on histopathological evaluation, which is subject to variability. While immunohistochemical staining (IHC) assists in distinguishing benign from malignant tissue, it involves increased work, higher costs, and diagnostic delays. Artificial intelligence (AI) presents a promising solution to reduce reliance on IHC by accurately classifying atypical glands and borderline morphologies in hematoxylin & eosin (H&E) stained tissue sections. In this study, we evaluated an AI model's ability to minimize IHC use without compromising diagnostic accuracy by retrospectively analyzing prostate core needle biopsies from routine diagnostics at three different pathology sites. These cohorts were composed exclusively of difficult cases where the diagnosing pathologists required IHC to finalize the diagnosis. The AI model demonstrated area under the curve values of 0.951-0.993 for detecting cancer in routine H&E-stained slides. Applying sensitivity-prioritized diagnostic thresholds reduced the need for IHC staining by 44.4%, 42.0%, and 20.7% in the three cohorts investigated, without a single false negative prediction. This AI model shows potential for optimizing IHC use, streamlining decision-making in prostate pathology, and alleviating resource burdens.
The impact of tissue detection on diagnostic artificial intelligence algorithms in digital pathology
Mar 29, 2025Abstract:Tissue detection is a crucial first step in most digital pathology applications. Details of the segmentation algorithm are rarely reported, and there is a lack of studies investigating the downstream effects of a poor segmentation algorithm. Disregarding tissue detection quality could create a bottleneck for downstream performance and jeopardize patient safety if diagnostically relevant parts of the specimen are excluded from analysis in clinical applications. This study aims to determine whether performance of downstream tasks is sensitive to the tissue detection method, and to compare performance of classical and AI-based tissue detection. To this end, we trained an AI model for Gleason grading of prostate cancer in whole slide images (WSIs) using two different tissue detection algorithms: thresholding (classical) and UNet++ (AI). A total of 33,823 WSIs scanned on five digital pathology scanners were used to train the tissue detection AI model. The downstream Gleason grading algorithm was trained and tested using 70,524 WSIs from 13 clinical sites scanned on 13 different scanners. There was a decrease from 116 (0.43%) to 22 (0.08%) fully undetected tissue samples when switching from thresholding-based tissue detection to AI-based, suggesting an AI model may be more reliable than a classical model for avoiding total failures on slides with unusual appearance. On the slides where tissue could be detected by both algorithms, no significant difference in overall Gleason grading performance was observed. However, tissue detection dependent clinically significant variations in AI grading were observed in 3.5% of malignant slides, highlighting the importance of robust tissue detection for optimal clinical performance of diagnostic AI.
Foundation Models -- A Panacea for Artificial Intelligence in Pathology?
Feb 28, 2025
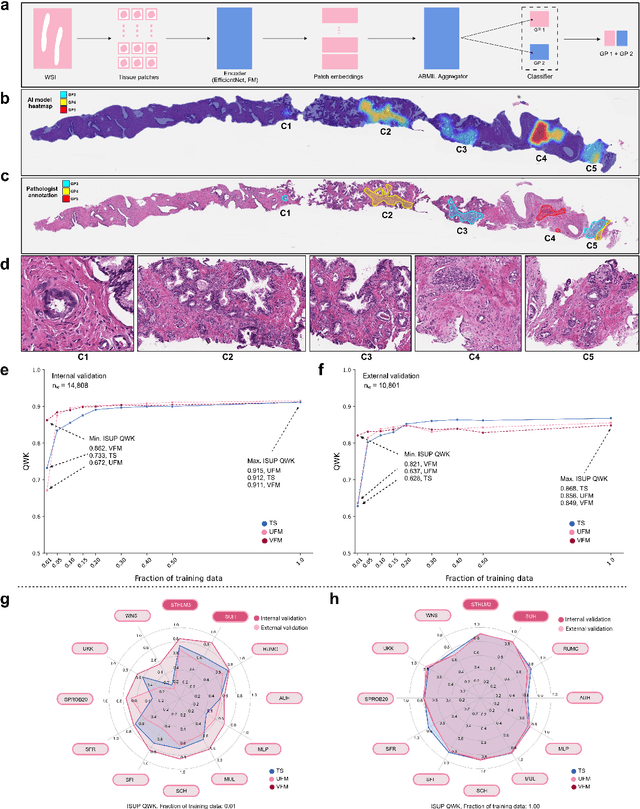

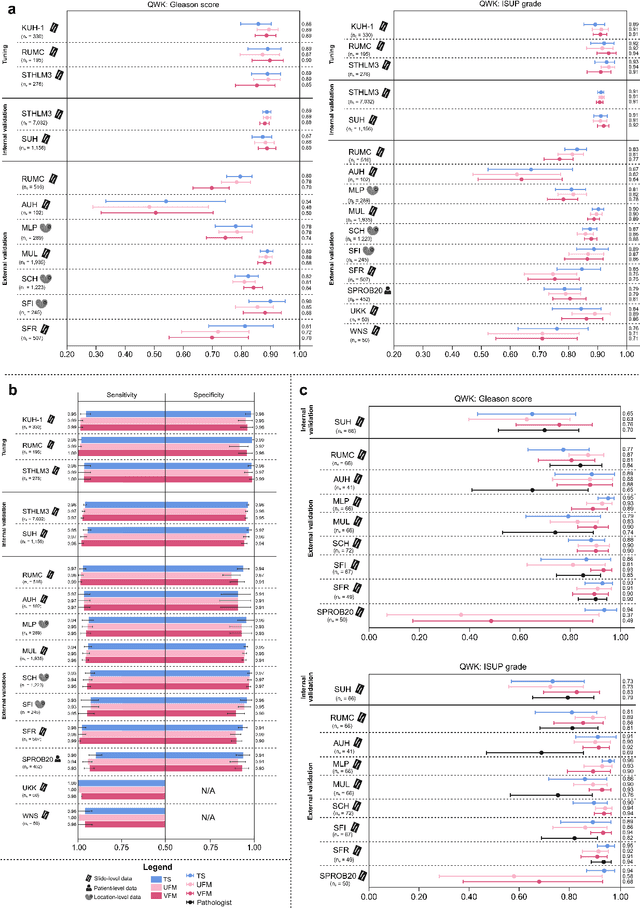
Abstract:The role of artificial intelligence (AI) in pathology has evolved from aiding diagnostics to uncovering predictive morphological patterns in whole slide images (WSIs). Recently, foundation models (FMs) leveraging self-supervised pre-training have been widely advocated as a universal solution for diverse downstream tasks. However, open questions remain about their clinical applicability and generalization advantages over end-to-end learning using task-specific (TS) models. Here, we focused on AI with clinical-grade performance for prostate cancer diagnosis and Gleason grading. We present the largest validation of AI for this task, using over 100,000 core needle biopsies from 7,342 patients across 15 sites in 11 countries. We compared two FMs with a fully end-to-end TS model in a multiple instance learning framework. Our findings challenge assumptions that FMs universally outperform TS models. While FMs demonstrated utility in data-scarce scenarios, their performance converged with - and was in some cases surpassed by - TS models when sufficient labeled training data were available. Notably, extensive task-specific training markedly reduced clinically significant misgrading, misdiagnosis of challenging morphologies, and variability across different WSI scanners. Additionally, FMs used up to 35 times more energy than the TS model, raising concerns about their sustainability. Our results underscore that while FMs offer clear advantages for rapid prototyping and research, their role as a universal solution for clinically applicable medical AI remains uncertain. For high-stakes clinical applications, rigorous validation and consideration of task-specific training remain critically important. We advocate for integrating the strengths of FMs and end-to-end learning to achieve robust and resource-efficient AI pathology solutions fit for clinical use.
Physical Color Calibration of Digital Pathology Scanners for Robust Artificial Intelligence Assisted Cancer Diagnosis
Jul 07, 2023



Abstract:The potential of artificial intelligence (AI) in digital pathology is limited by technical inconsistencies in the production of whole slide images (WSIs), leading to degraded AI performance and posing a challenge for widespread clinical application as fine-tuning algorithms for each new site is impractical. Changes in the imaging workflow can also lead to compromised diagnoses and patient safety risks. We evaluated whether physical color calibration of scanners can standardize WSI appearance and enable robust AI performance. We employed a color calibration slide in four different laboratories and evaluated its impact on the performance of an AI system for prostate cancer diagnosis on 1,161 WSIs. Color standardization resulted in consistently improved AI model calibration and significant improvements in Gleason grading performance. The study demonstrates that physical color calibration provides a potential solution to the variation introduced by different scanners, making AI-based cancer diagnostics more reliable and applicable in clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge