Anastasios N Angelopoulos
UC Berkeley
Automatically Adaptive Conformal Risk Control
Jun 25, 2024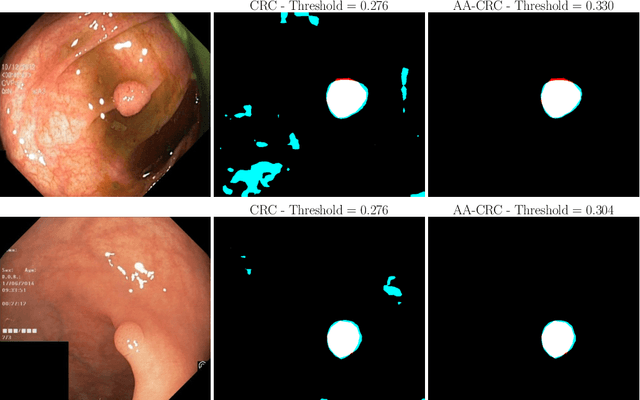

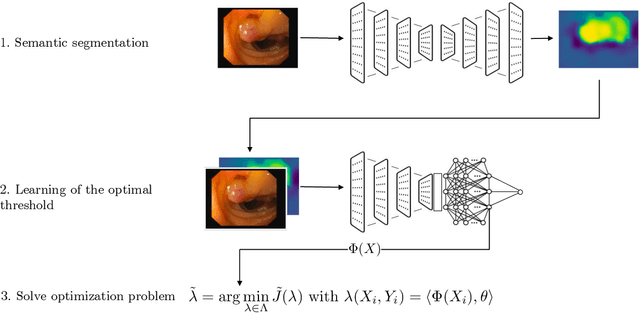
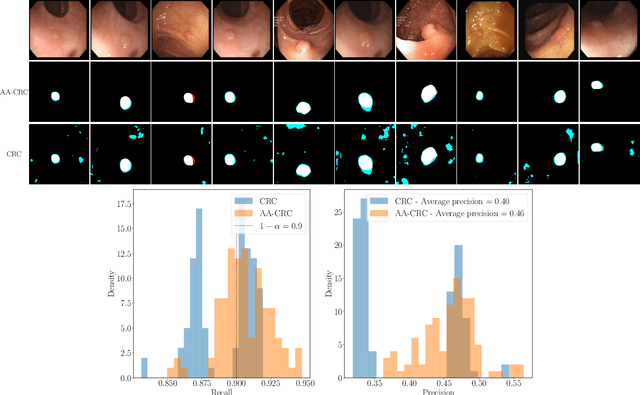
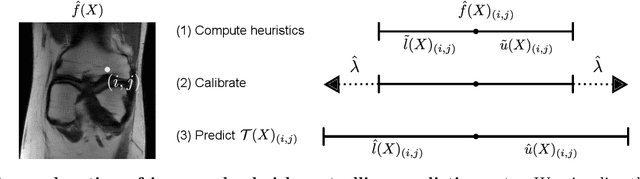
Abstract:Science and technology have a growing need for effective mechanisms that ensure reliable, controlled performance from black-box machine learning algorithms. These performance guarantees should ideally hold conditionally on the input-that is the performance guarantees should hold, at least approximately, no matter what the input. However, beyond stylized discrete groupings such as ethnicity and gender, the right notion of conditioning can be difficult to define. For example, in problems such as image segmentation, we want the uncertainty to reflect the intrinsic difficulty of the test sample, but this may be difficult to capture via a conditioning event. Building on the recent work of Gibbs et al. [2023], we propose a methodology for achieving approximate conditional control of statistical risks-the expected value of loss functions-by adapting to the difficulty of test samples. Our framework goes beyond traditional conditional risk control based on user-provided conditioning events to the algorithmic, data-driven determination of appropriate function classes for conditioning. We apply this framework to various regression and segmentation tasks, enabling finer-grained control over model performance and demonstrating that by continuously monitoring and adjusting these parameters, we can achieve superior precision compared to conventional risk-control methods.
Image-to-Image Regression with Distribution-Free Uncertainty Quantification and Applications in Imaging
Feb 10, 2022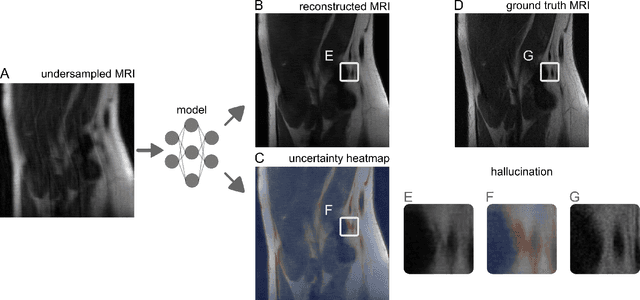
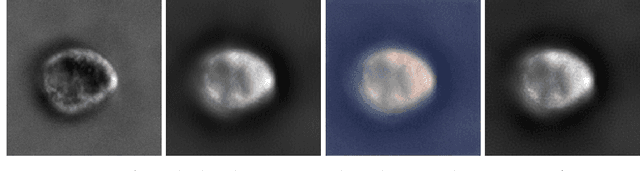
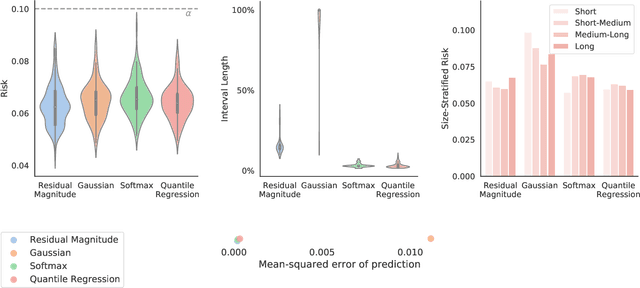
Abstract:Image-to-image regression is an important learning task, used frequently in biological imaging. Current algorithms, however, do not generally offer statistical guarantees that protect against a model's mistakes and hallucinations. To address this, we develop uncertainty quantification techniques with rigorous statistical guarantees for image-to-image regression problems. In particular, we show how to derive uncertainty intervals around each pixel that are guaranteed to contain the true value with a user-specified confidence probability. Our methods work in conjunction with any base machine learning model, such as a neural network, and endow it with formal mathematical guarantees -- regardless of the true unknown data distribution or choice of model. Furthermore, they are simple to implement and computationally inexpensive. We evaluate our procedure on three image-to-image regression tasks: quantitative phase microscopy, accelerated magnetic resonance imaging, and super-resolution transmission electron microscopy of a Drosophila melanogaster brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge