Amir Manbachi
Convolutional Deep Operator Networks for Learning Nonlinear Focused Ultrasound Wave Propagation in Heterogeneous Spinal Cord Anatomy
Dec 20, 2024



Abstract:Focused ultrasound (FUS) therapy is a promising tool for optimally targeted treatment of spinal cord injuries (SCI), offering submillimeter precision to enhance blood flow at injury sites while minimizing impact on surrounding tissues. However, its efficacy is highly sensitive to the placement of the ultrasound source, as the spinal cord's complex geometry and acoustic heterogeneity distort and attenuate the FUS signal. Current approaches rely on computer simulations to solve the governing wave propagation equations and compute patient-specific pressure maps using ultrasound images of the spinal cord anatomy. While accurate, these high-fidelity simulations are computationally intensive, taking up to hours to complete parameter sweeps, which is impractical for real-time surgical decision-making. To address this bottleneck, we propose a convolutional deep operator network (DeepONet) to rapidly predict FUS pressure fields in patient spinal cords. Unlike conventional neural networks, DeepONets are well equipped to approximate the solution operator of the parametric partial differential equations (PDEs) that govern the behavior of FUS waves with varying initial and boundary conditions (i.e., new transducer locations or spinal cord geometries) without requiring extensive simulations. Trained on simulated pressure maps across diverse patient anatomies, this surrogate model achieves real-time predictions with only a 2% loss on the test set, significantly accelerating the modeling of nonlinear physical systems in heterogeneous domains. By facilitating rapid parameter sweeps in surgical settings, this work provides a crucial step toward precise and individualized solutions in neurosurgical treatments.
A novel open-source ultrasound dataset with deep learning benchmarks for spinal cord injury localization and anatomical segmentation
Sep 24, 2024



Abstract:While deep learning has catalyzed breakthroughs across numerous domains, its broader adoption in clinical settings is inhibited by the costly and time-intensive nature of data acquisition and annotation. To further facilitate medical machine learning, we present an ultrasound dataset of 10,223 Brightness-mode (B-mode) images consisting of sagittal slices of porcine spinal cords (N=25) before and after a contusion injury. We additionally benchmark the performance metrics of several state-of-the-art object detection algorithms to localize the site of injury and semantic segmentation models to label the anatomy for comparison and creation of task-specific architectures. Finally, we evaluate the zero-shot generalization capabilities of the segmentation models on human ultrasound spinal cord images to determine whether training on our porcine dataset is sufficient for accurately interpreting human data. Our results show that the YOLOv8 detection model outperforms all evaluated models for injury localization, achieving a mean Average Precision (mAP50-95) score of 0.606. Segmentation metrics indicate that the DeepLabv3 segmentation model achieves the highest accuracy on unseen porcine anatomy, with a Mean Dice score of 0.587, while SAMed achieves the highest Mean Dice score generalizing to human anatomy (0.445). To the best of our knowledge, this is the largest annotated dataset of spinal cord ultrasound images made publicly available to researchers and medical professionals, as well as the first public report of object detection and segmentation architectures to assess anatomical markers in the spinal cord for methodology development and clinical applications.
Unidirectional brain-computer interface: Artificial neural network encoding natural images to fMRI response in the visual cortex
Sep 26, 2023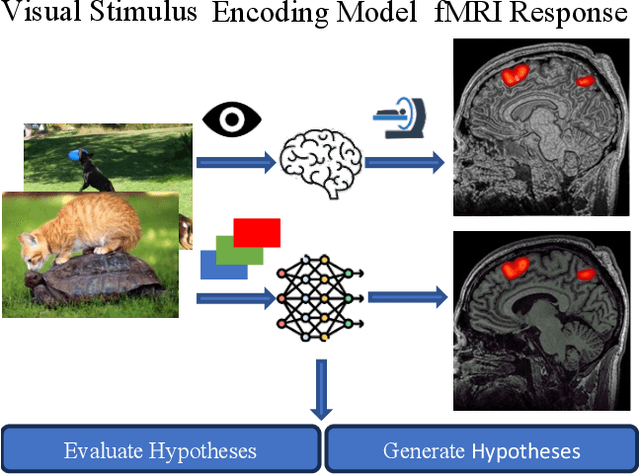

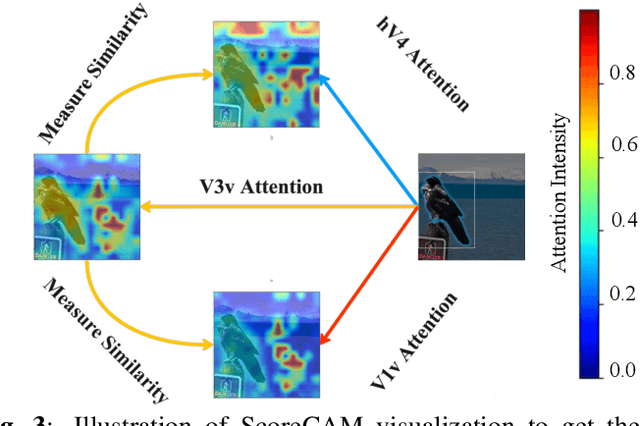
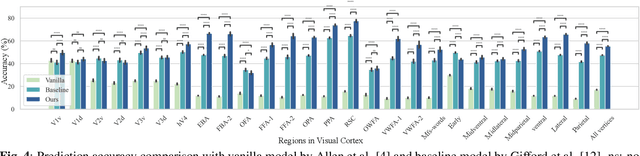
Abstract:While significant advancements in artificial intelligence (AI) have catalyzed progress across various domains, its full potential in understanding visual perception remains underexplored. We propose an artificial neural network dubbed VISION, an acronym for "Visual Interface System for Imaging Output of Neural activity," to mimic the human brain and show how it can foster neuroscientific inquiries. Using visual and contextual inputs, this multimodal model predicts the brain's functional magnetic resonance imaging (fMRI) scan response to natural images. VISION successfully predicts human hemodynamic responses as fMRI voxel values to visual inputs with an accuracy exceeding state-of-the-art performance by 45%. We further probe the trained networks to reveal representational biases in different visual areas, generate experimentally testable hypotheses, and formulate an interpretable metric to associate these hypotheses with cortical functions. With both a model and evaluation metric, the cost and time burdens associated with designing and implementing functional analysis on the visual cortex could be reduced. Our work suggests that the evolution of computational models may shed light on our fundamental understanding of the visual cortex and provide a viable approach toward reliable brain-machine interfaces.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge