Alvaro Rodriguez
Toward a Theory of Causation for Interpreting Neural Code Models
Feb 07, 2023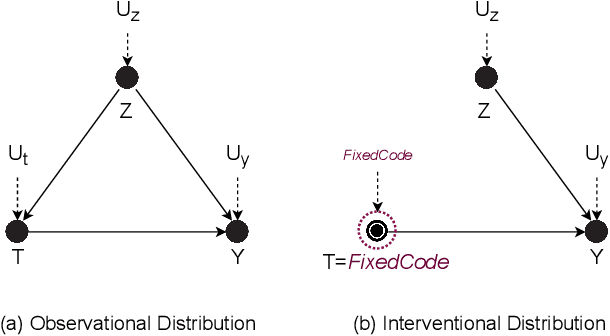
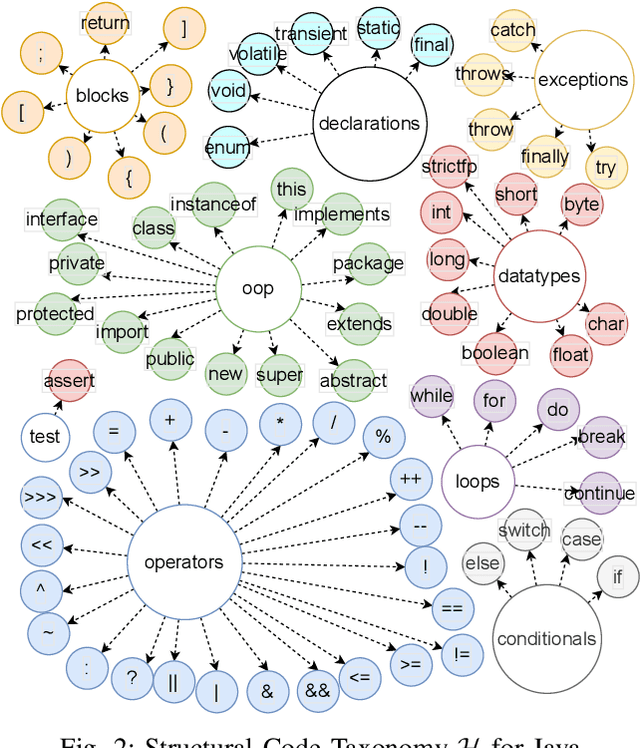
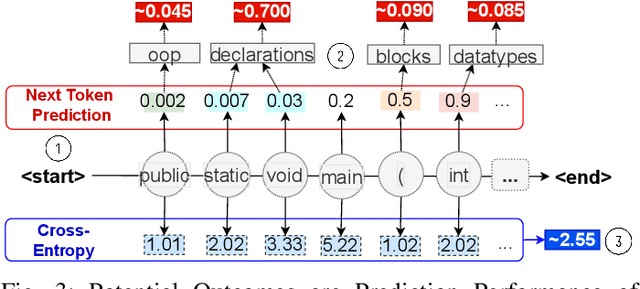
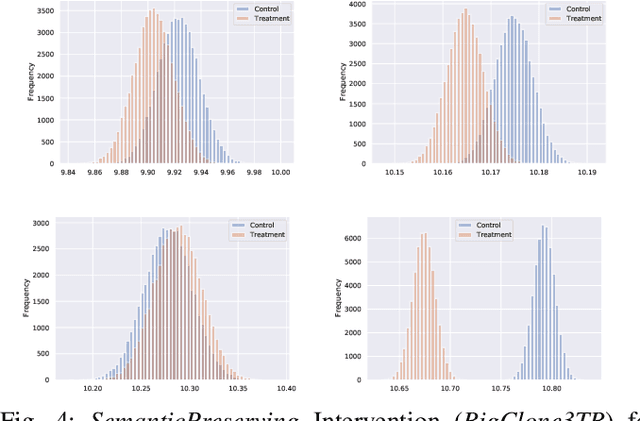
Abstract:Neural Language Models of Code, or Neural Code Models (NCMs), are rapidly progressing from research prototypes to commercial developer tools. As such, understanding the capabilities and limitations of such models is becoming critical. However, the abilities of these models are typically measured using automated metrics that often only reveal a portion of their real-world performance. While, in general, the performance of NCMs appears promising, currently much is unknown about how such models arrive at decisions. To this end, this paper introduces $do_{code}$, a post-hoc interpretability methodology specific to NCMs that is capable of explaining model predictions. $do_{code}$ is based upon causal inference to enable programming language-oriented explanations. While the theoretical underpinnings of $do_{code}$ are extensible to exploring different model properties, we provide a concrete instantiation that aims to mitigate the impact of spurious correlations by grounding explanations of model behavior in properties of programming languages. To demonstrate the practical benefit of $do_{code}$, we illustrate the insights that our framework can provide by performing a case study on two popular deep learning architectures and nine NCMs. The results of this case study illustrate that our studied NCMs are sensitive to changes in code syntax and statistically learn to predict tokens related to blocks of code (e.g., brackets, parenthesis, semicolon) with less confounding bias as compared to other programming language constructs. These insights demonstrate the potential of $do_{code}$ as a useful model debugging mechanism that may aid in discovering biases and limitations in NCMs.
UmUTracker: A versatile MATLAB program for automated particle tracking of 2D light microscopy or 3D digital holography data
Apr 21, 2017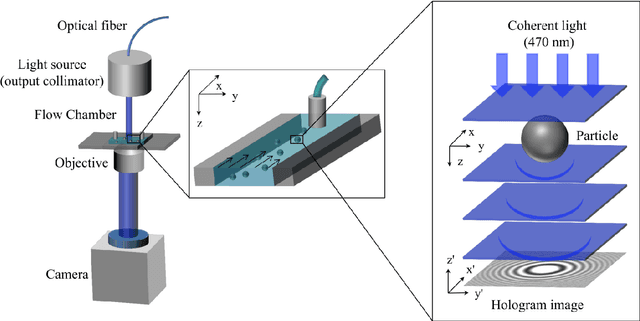
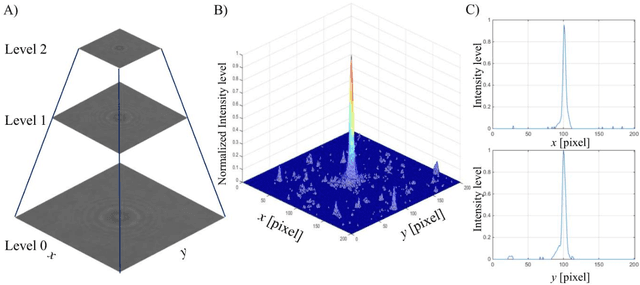
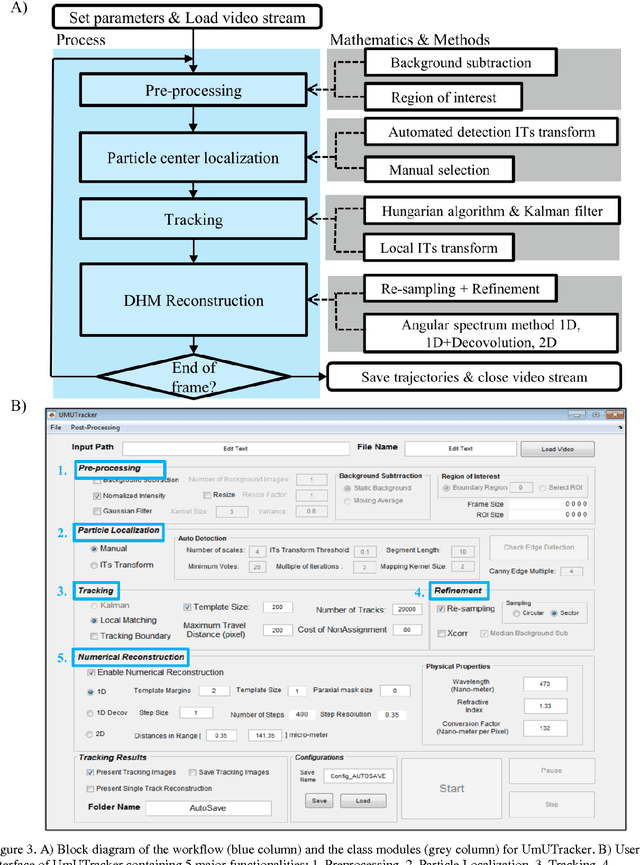
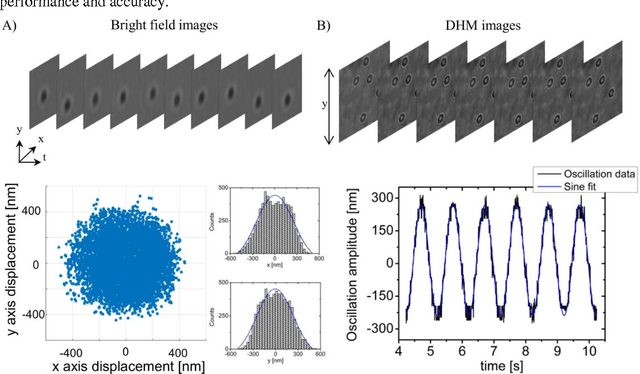
Abstract:We present a versatile and fast MATLAB program (UmUTracker) that automatically detects and tracks particles by analyzing video sequences acquired by either light microscopy or digital in-line holographic microscopy. Our program detects the 2D lateral positions of particles with an algorithm based on the isosceles triangle transform, and reconstructs their 3D axial positions by a fast implementation of the Rayleigh-Sommerfeld model using a radial intensity profile. To validate the accuracy and performance of our program, we first track the 2D position of polystyrene particles using bright field and digital holographic microscopy. Second, we determine the 3D particle position by analyzing synthetic and experimentally acquired holograms. Finally, to highlight the full program features, we profile the microfluidic flow in a 100 micrometer high flow chamber. This result agrees with computational fluid dynamic simulations. On a regular desktop computer UmUTracker can detect, analyze, and track multiple particles at 5 frames per second for a template size of 201 x 201 in a 1024 x 1024 image. To enhance usability and to make it easy to implement new functions we used object-oriented programming. UmUTracker is suitable for studies related to: particle dynamics, cell localization, colloids and microfluidic flow measurement.
A robust particle detection algorithm based on symmetry
May 11, 2016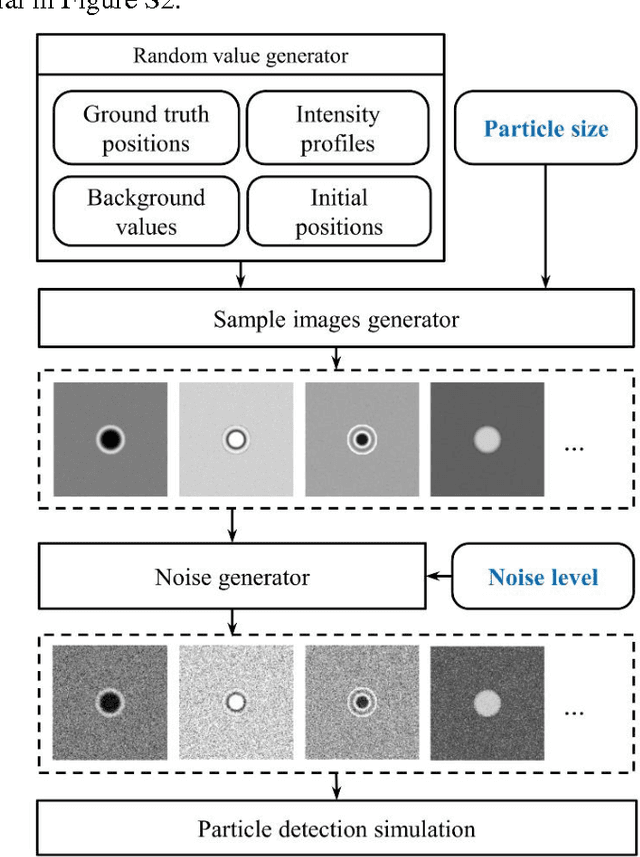
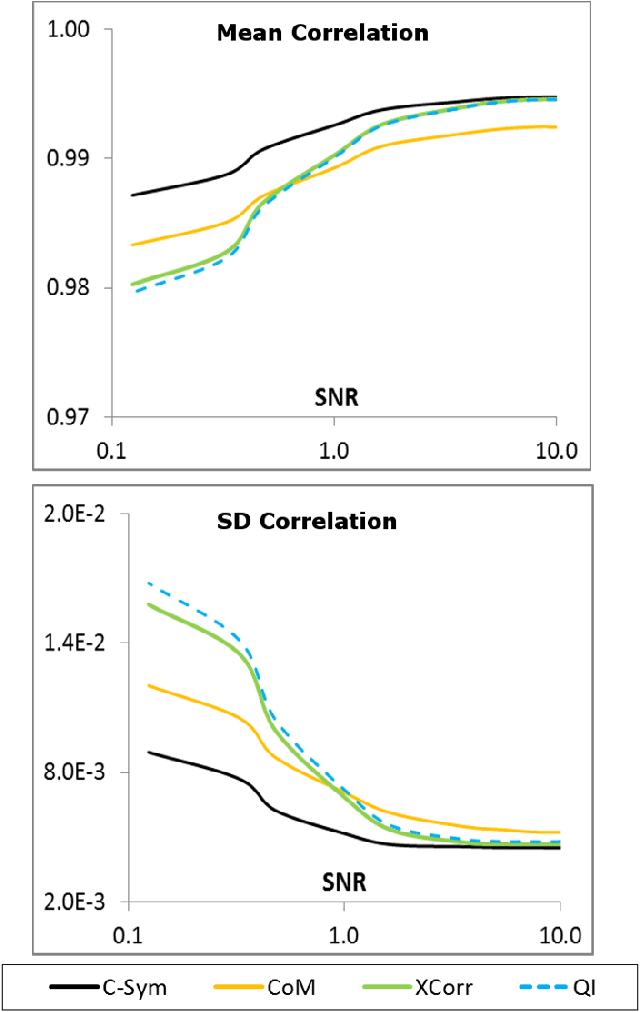
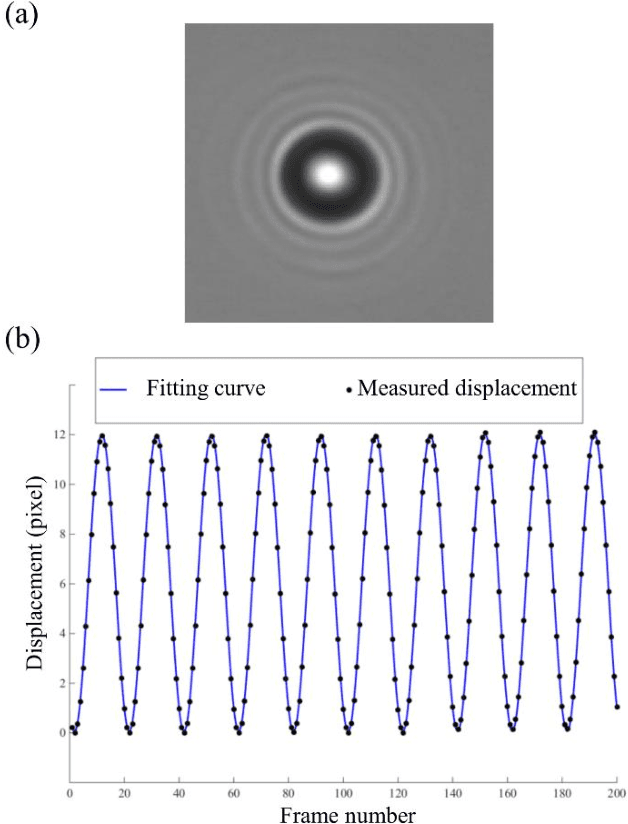
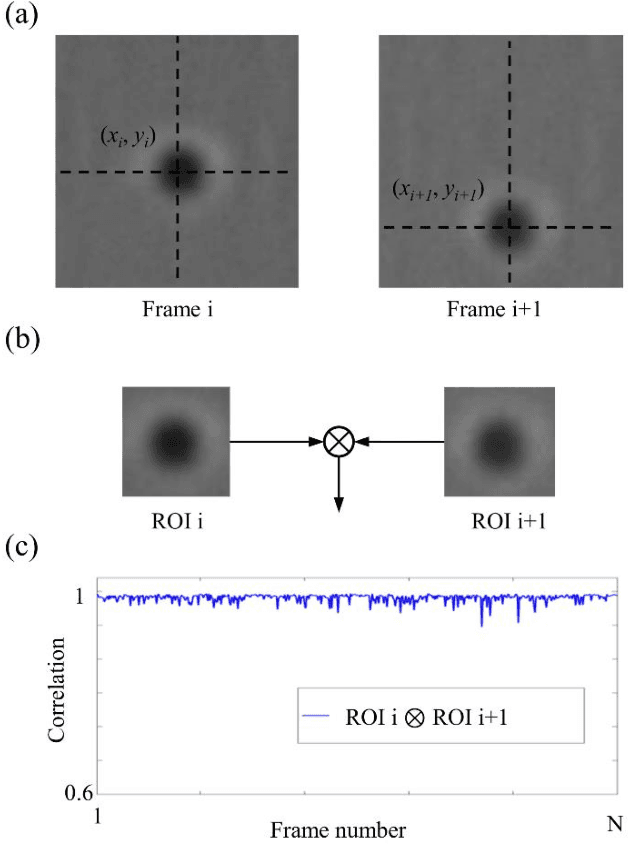
Abstract:Particle tracking is common in many biophysical, ecological, and micro-fluidic applications. Reliable tracking information is heavily dependent on of the system under study and algorithms that correctly determines particle position between images. However, in a real environmental context with the presence of noise including particular or dissolved matter in water, and low and fluctuating light conditions, many algorithms fail to obtain reliable information. We propose a new algorithm, the Circular Symmetry algorithm (C-Sym), for detecting the position of a circular particle with high accuracy and precision in noisy conditions. The algorithm takes advantage of the spatial symmetry of the particle allowing for subpixel accuracy. We compare the proposed algorithm with four different methods using both synthetic and experimental datasets. The results show that C-Sym is the most accurate and precise algorithm when tracking micro-particles in all tested conditions and it has the potential for use in applications including tracking biota in their environment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge