Alper Yegenoglu
Multi-Agent Systems Powered by Large Language Models: Applications in Swarm Intelligence
Mar 05, 2025Abstract:This work examines the integration of large language models (LLMs) into multi-agent simulations by replacing the hard-coded programs of agents with LLM-driven prompts. The proposed approach is showcased in the context of two examples of complex systems from the field of swarm intelligence: ant colony foraging and bird flocking. Central to this study is a toolchain that integrates LLMs with the NetLogo simulation platform, leveraging its Python extension to enable communication with GPT-4o via the OpenAI API. This toolchain facilitates prompt-driven behavior generation, allowing agents to respond adaptively to environmental data. For both example applications mentioned above, we employ both structured, rule-based prompts and autonomous, knowledge-driven prompts. Our work demonstrates how this toolchain enables LLMs to study self-organizing processes and induce emergent behaviors within multi-agent environments, paving the way for new approaches to exploring intelligent systems and modeling swarm intelligence inspired by natural phenomena. We provide the code, including simulation files and data at https://github.com/crjimene/swarm_gpt.
Two-compartment neuronal spiking model expressing brain-state specific apical-amplification, -isolation and -drive regimes
Nov 10, 2023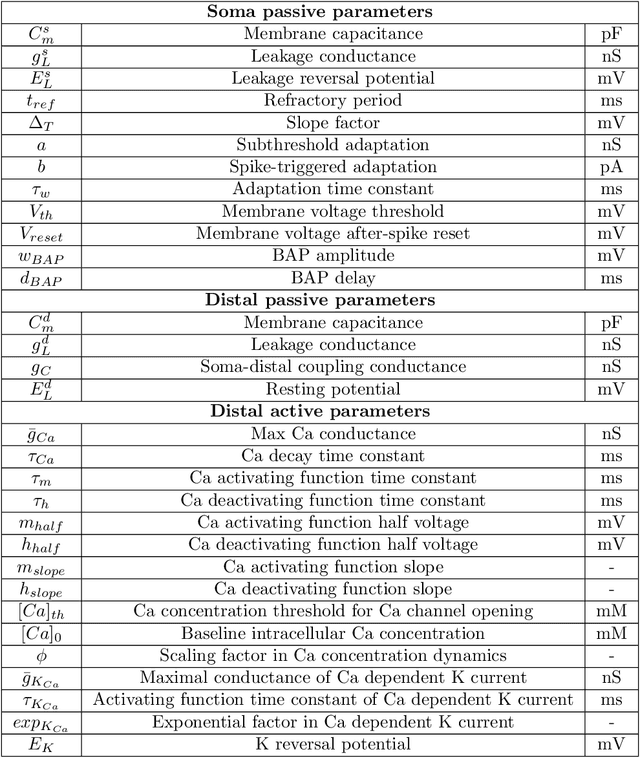
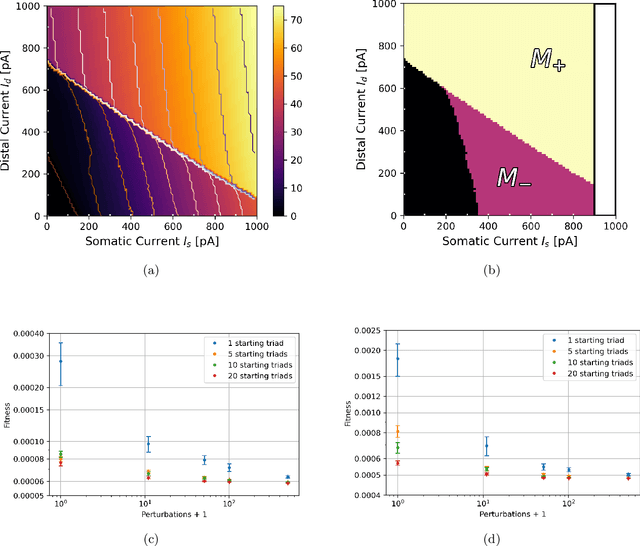
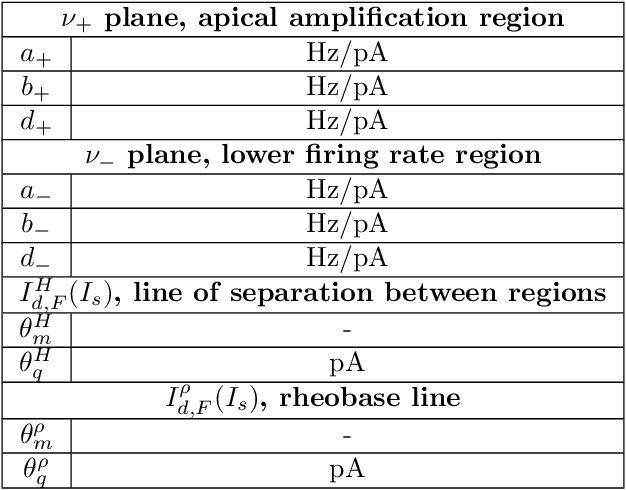
Abstract:There is mounting experimental evidence that brain-state specific neural mechanisms supported by connectomic architectures serve to combine past and contextual knowledge with current, incoming flow of evidence (e.g. from sensory systems). Such mechanisms are distributed across multiple spatial and temporal scales and require dedicated support at the levels of individual neurons and synapses. A prominent feature in the neocortex is the structure of large, deep pyramidal neurons which show a peculiar separation between an apical dendritic compartment and a basal dentritic/peri-somatic compartment, with distinctive patterns of incoming connections and brain-state specific activation mechanisms, namely apical-amplification, -isolation and -drive associated to the wakefulness, deeper NREM sleep stages and REM sleep. The cognitive roles of apical mechanisms have been demonstrated in behaving animals. In contrast, classical models of learning spiking networks are based on single compartment neurons that miss the description of mechanisms to combine apical and basal/somatic information. This work aims to provide the computational community with a two-compartment spiking neuron model which includes features that are essential for supporting brain-state specific learning and with a piece-wise linear transfer function (ThetaPlanes) at highest abstraction level to be used in large scale bio-inspired artificial intelligence systems. A machine learning algorithm, constrained by a set of fitness functions, selected the parameters defining neurons expressing the desired apical mechanisms.
Emergent communication enhances foraging behaviour in evolved swarms controlled by Spiking Neural Networks
Dec 16, 2022Abstract:Social insects such as ants communicate via pheromones which allows them to coordinate their activity and solve complex tasks as a swarm, e.g. foraging for food. This behaviour was shaped through evolutionary processes. In computational models, self-coordination in swarms has been implemented using probabilistic or action rules to shape the decision of each agent and the collective behaviour. However, manual tuned decision rules may limit the behaviour of the swarm. In this work we investigate the emergence of self-coordination and communication in evolved swarms without defining any rule. We evolve a swarm of agents representing an ant colony. We use a genetic algorithm to optimize a spiking neural network (SNN) which serves as an artificial brain to control the behaviour of each agent. The goal of the colony is to find optimal ways to forage for food in the shortest amount of time. In the evolutionary phase, the ants are able to learn to collaborate by depositing pheromone near food piles and near the nest to guide its cohorts. The pheromone usage is not encoded into the network; instead, this behaviour is established through the optimization procedure. We observe that pheromone-based communication enables the ants to perform better in comparison to colonies where communication did not emerge. We assess the foraging performance by comparing the SNN based model to a rule based system. Our results show that the SNN based model can complete the foraging task more efficiently in a shorter time. Our approach illustrates that even in the absence of pre-defined rules, self coordination via pheromone emerges as a result of the network optimization. This work serves as a proof of concept for the possibility of creating complex applications utilizing SNNs as underlying architectures for multi-agent interactions where communication and self-coordination is desired.
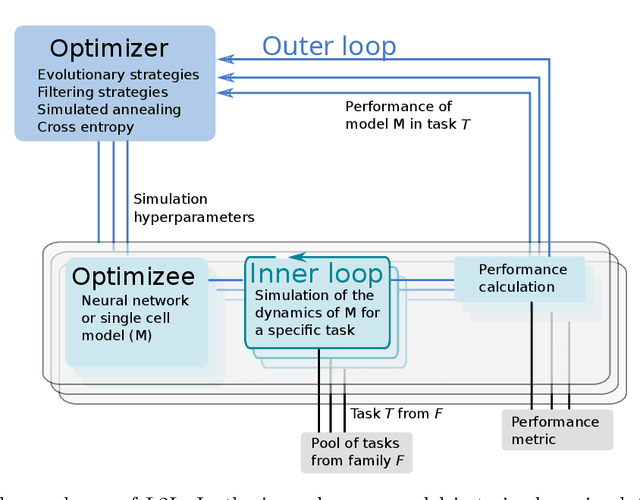
Exploring hyper-parameter spaces of neuroscience models on high performance computers with Learning to Learn
Feb 28, 2022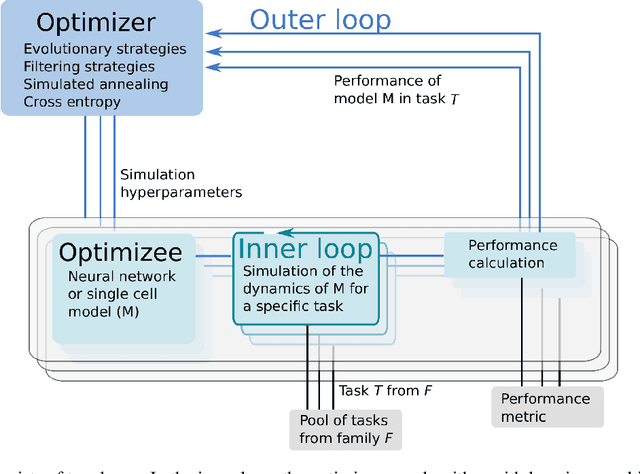
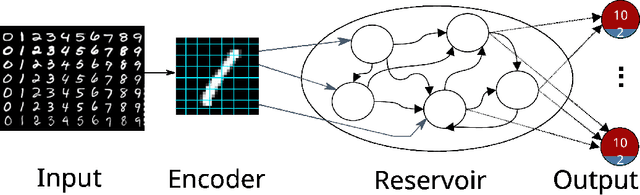
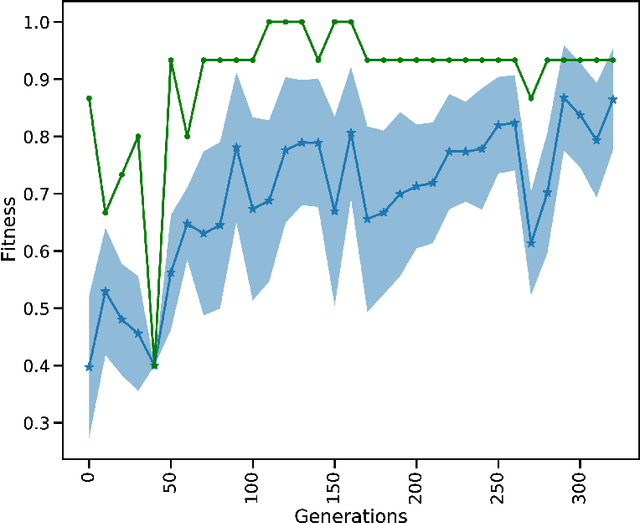
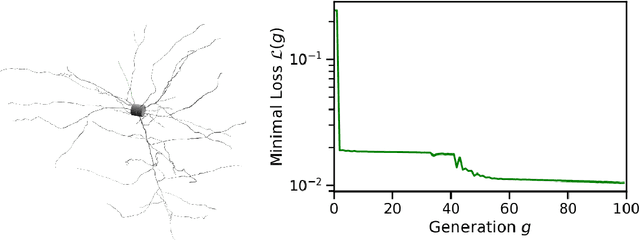
Abstract:Neuroscience models commonly have a high number of degrees of freedom and only specific regions within the parameter space are able to produce dynamics of interest. This makes the development of tools and strategies to efficiently find these regions of high importance to advance brain research. Exploring the high dimensional parameter space using numerical simulations has been a frequently used technique in the last years in many areas of computational neuroscience. High performance computing (HPC) can provide today a powerful infrastructure to speed up explorations and increase our general understanding of the model's behavior in reasonable times.
Generalised learning of time-series: Ornstein-Uhlenbeck processes
Oct 21, 2019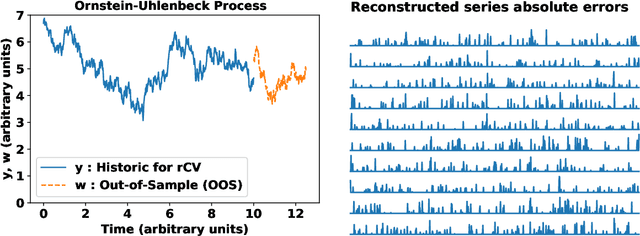
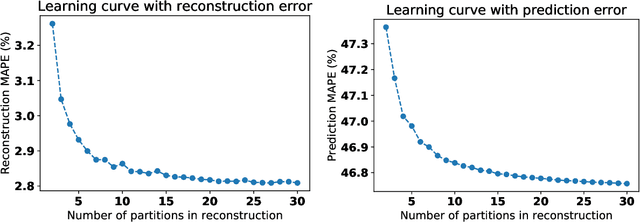
Abstract:In machine learning, statistics, econometrics and statistical physics, $k$-fold cross-validation (CV) is used as a standard approach in quantifying the generalization performance of a statistical model. Applying this approach directly to time series models is avoided by practitioners due to intrinsic nature of serial correlations in the ordered data due to implications like absurdity of using future data to predict past and non-stationarity issues. In this work, we propose a technique called {\it reconstructive cross validation} ($rCV$) that avoids all these issues enabling generalized learning in time-series as a meta-algorithm. In $rCV$, data points in the test fold, randomly selected points from the time series, are first removed. Then, a secondary time series model or a technique is used in reconstructing the removed points from the test fold, i.e., imputation or smoothing. Thereafter, the primary model is build using new dataset coming from the secondary model or a technique. The performance of the primary model on the test set by computing the deviations from the originally removed and out-of-sample (OSS) data are evaluated simultaneously. This amounts to reconstruction and prediction errors. By this procedure serial correlations and data order is retained and $k$-fold cross-validation is reached generically. If reconstruction model uses a technique whereby the existing data points retained exactly, such as Gaussian process regression, the reconstruction itself will not result in information loss from non-reconstructed portion of the original data points. We have applied $rCV$ to estimate the general performance of the model build on simulated Ornstein-Uhlenbeck process. We have shown an approach to build a time-series learning curves utilizing $rCV$.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge