Aline Sindel
Pattern Recognition Lab, FAU Erlangen-Nürnberg
Coarse-to-Fine Non-rigid Multi-modal Image Registration for Historical Panel Paintings based on Crack Structures
Jan 22, 2026Abstract:Art technological investigations of historical panel paintings rely on acquiring multi-modal image data, including visual light photography, infrared reflectography, ultraviolet fluorescence photography, x-radiography, and macro photography. For a comprehensive analysis, the multi-modal images require pixel-wise alignment, which is still often performed manually. Multi-modal image registration can reduce this laborious manual work, is substantially faster, and enables higher precision. Due to varying image resolutions, huge image sizes, non-rigid distortions, and modality-dependent image content, registration is challenging. Therefore, we propose a coarse-to-fine non-rigid multi-modal registration method efficiently relying on sparse keypoints and thin-plate-splines. Historical paintings exhibit a fine crack pattern, called craquelure, on the paint layer, which is captured by all image systems and is well-suited as a feature for registration. In our one-stage non-rigid registration approach, we employ a convolutional neural network for joint keypoint detection and description based on the craquelure and a graph neural network for descriptor matching in a patch-based manner, and filter matches based on homography reprojection errors in local areas. For coarse-to-fine registration, we introduce a novel multi-level keypoint refinement approach to register mixed-resolution images up to the highest resolution. We created a multi-modal dataset of panel paintings with a high number of keypoint annotations, and a large test set comprising five multi-modal domains and varying image resolutions. The ablation study demonstrates the effectiveness of all modules of our refinement method. Our proposed approaches achieve the best registration results compared to competing keypoint and dense matching methods and refinement methods.
Data Augmentation via Latent Diffusion Models for Detecting Smell-Related Objects in Historical Artworks
Sep 18, 2025Abstract:Finding smell references in historic artworks is a challenging problem. Beyond artwork-specific challenges such as stylistic variations, their recognition demands exceptionally detailed annotation classes, resulting in annotation sparsity and extreme class imbalance. In this work, we explore the potential of synthetic data generation to alleviate these issues and enable accurate detection of smell-related objects. We evaluate several diffusion-based augmentation strategies and demonstrate that incorporating synthetic data into model training can improve detection performance. Our findings suggest that leveraging the large-scale pretraining of diffusion models offers a promising approach for improving detection accuracy, particularly in niche applications where annotations are scarce and costly to obtain. Furthermore, the proposed approach proves to be effective even with relatively small amounts of data, and scaling it up provides high potential for further enhancements.
Automated Segmentation and Analysis of Cone Photoreceptors in Multimodal Adaptive Optics Imaging
Oct 19, 2024Abstract:Accurate detection and segmentation of cone cells in the retina are essential for diagnosing and managing retinal diseases. In this study, we used advanced imaging techniques, including confocal and non-confocal split detector images from adaptive optics scanning light ophthalmoscopy (AOSLO), to analyze photoreceptors for improved accuracy. Precise segmentation is crucial for understanding each cone cell's shape, area, and distribution. It helps to estimate the surrounding areas occupied by rods, which allows the calculation of the density of cone photoreceptors in the area of interest. In turn, density is critical for evaluating overall retinal health and functionality. We explored two U-Net-based segmentation models: StarDist for confocal and Cellpose for calculated modalities. Analyzing cone cells in images from two modalities and achieving consistent results demonstrates the study's reliability and potential for clinical application.
Generalist Segmentation Algorithm for Photoreceptors Analysis in Adaptive Optics Imaging
Aug 27, 2024Abstract:Analyzing the cone photoreceptor pattern in images obtained from the living human retina using quantitative methods can be crucial for the early detection and management of various eye conditions. Confocal adaptive optics scanning light ophthalmoscope (AOSLO) imaging enables visualization of the cones from reflections of waveguiding cone photoreceptors. While there have been significant improvements in automated algorithms for segmenting cones in confocal AOSLO images, the process of labelling data remains labor-intensive and manual. This paper introduces a method based on deep learning (DL) for detecting and segmenting cones in AOSLO images. The models were trained on a semi-automatically labelled dataset of 20 AOSLO batches of images of 18 participants for 0$^{\circ}$, 1$^{\circ}$, and 2$^{\circ}$ from the foveal center. F1 scores were 0.968, 0.958, and 0.954 for 0$^{\circ}$, 1$^{\circ}$, and 2$^{\circ}$, respectively, which is better than previously reported DL approaches. Our method minimizes the need for labelled data by only necessitating a fraction of labelled cones, which is especially beneficial in the field of ophthalmology, where labelled data can often be limited.
A Vessel-Segmentation-Based CycleGAN for Unpaired Multi-modal Retinal Image Synthesis
Jun 05, 2023Abstract:Unpaired image-to-image translation of retinal images can efficiently increase the training dataset for deep-learning-based multi-modal retinal registration methods. Our method integrates a vessel segmentation network into the image-to-image translation task by extending the CycleGAN framework. The segmentation network is inserted prior to a UNet vision transformer generator network and serves as a shared representation between both domains. We reformulate the original identity loss to learn the direct mapping between the vessel segmentation and the real image. Additionally, we add a segmentation loss term to ensure shared vessel locations between fake and real images. In the experiments, our method shows a visually realistic look and preserves the vessel structures, which is a prerequisite for generating multi-modal training data for image registration.
* Accepted to BVM 2023
ArtFacePoints: High-resolution Facial Landmark Detection in Paintings and Prints
Oct 17, 2022
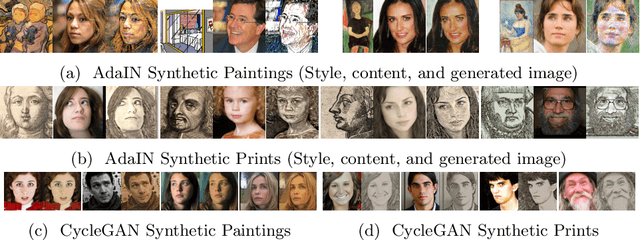
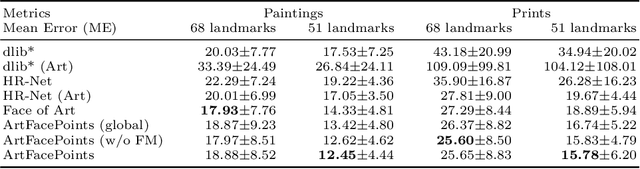
Abstract:Facial landmark detection plays an important role for the similarity analysis in artworks to compare portraits of the same or similar artists. With facial landmarks, portraits of different genres, such as paintings and prints, can be automatically aligned using control-point-based image registration. We propose a deep-learning-based method for facial landmark detection in high-resolution images of paintings and prints. It divides the task into a global network for coarse landmark prediction and multiple region networks for precise landmark refinement in regions of the eyes, nose, and mouth that are automatically determined based on the predicted global landmark coordinates. We created a synthetically augmented facial landmark art dataset including artistic style transfer and geometric landmark shifts. Our method demonstrates an accurate detection of the inner facial landmarks for our high-resolution dataset of artworks while being comparable for a public low-resolution artwork dataset in comparison to competing methods.
A Multi-modal Registration and Visualization Software Tool for Artworks using CraquelureNet
Aug 18, 2022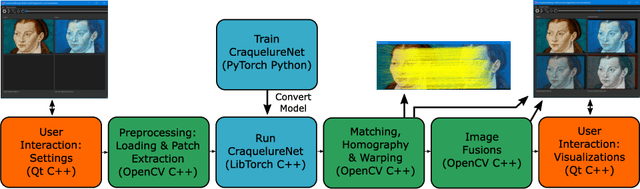
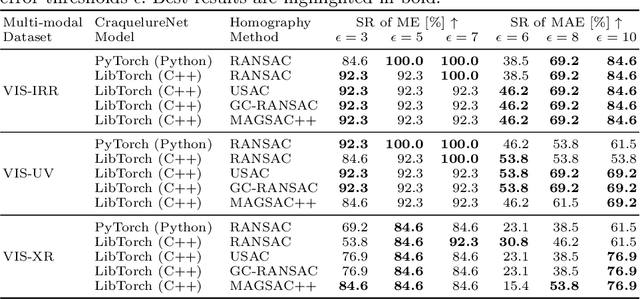
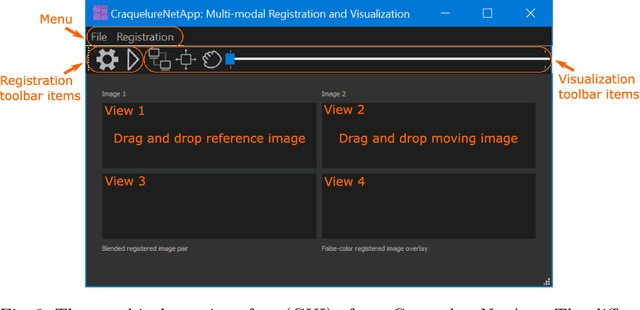
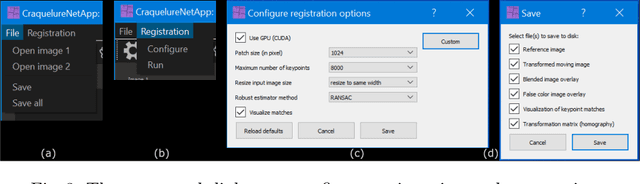
Abstract:For art investigations of paintings, multiple imaging technologies, such as visual light photography, infrared reflectography, ultraviolet fluorescence photography, and x-radiography are often used. For a pixel-wise comparison, the multi-modal images have to be registered. We present a registration and visualization software tool, that embeds a convolutional neural network to extract cross-modal features of the crack structures in historical paintings for automatic registration. The graphical user interface processes the user's input to configure the registration parameters and to interactively adapt the image views with the registered pair and image overlays, such as by individual or synchronized zoom or movements of the views. In the evaluation, we qualitatively and quantitatively show the effectiveness of our software tool in terms of registration performance and short inference time on multi-modal paintings and its transferability by applying our method to historical prints.
Multi-modal Retinal Image Registration Using a Keypoint-Based Vessel Structure Aligning Network
Jul 21, 2022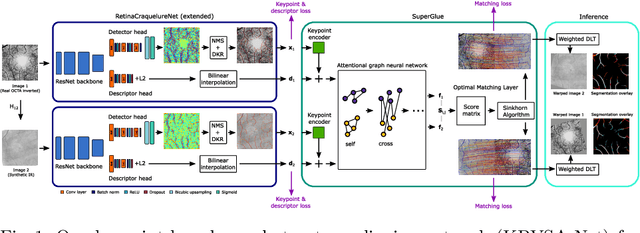
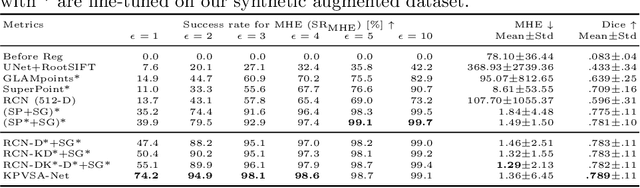
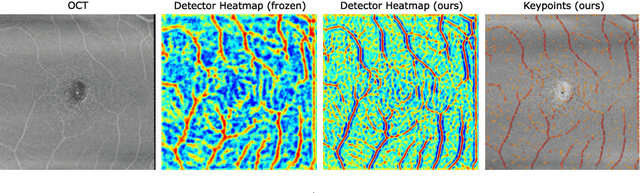
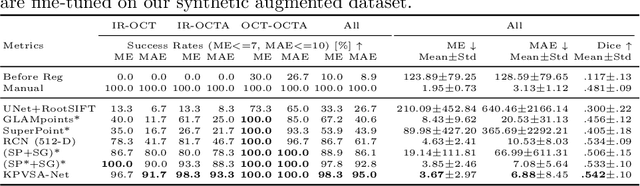
Abstract:In ophthalmological imaging, multiple imaging systems, such as color fundus, infrared, fluorescein angiography, optical coherence tomography (OCT) or OCT angiography, are often involved to make a diagnosis of retinal disease. Multi-modal retinal registration techniques can assist ophthalmologists by providing a pixel-based comparison of aligned vessel structures in images from different modalities or acquisition times. To this end, we propose an end-to-end trainable deep learning method for multi-modal retinal image registration. Our method extracts convolutional features from the vessel structure for keypoint detection and description and uses a graph neural network for feature matching. The keypoint detection and description network and graph neural network are jointly trained in a self-supervised manner using synthetic multi-modal image pairs and are guided by synthetically sampled ground truth homographies. Our method demonstrates higher registration accuracy as competing methods for our synthetic retinal dataset and generalizes well for our real macula dataset and a public fundus dataset.
SliTraNet: Automatic Detection of Slide Transitions in Lecture Videos using Convolutional Neural Networks
Feb 07, 2022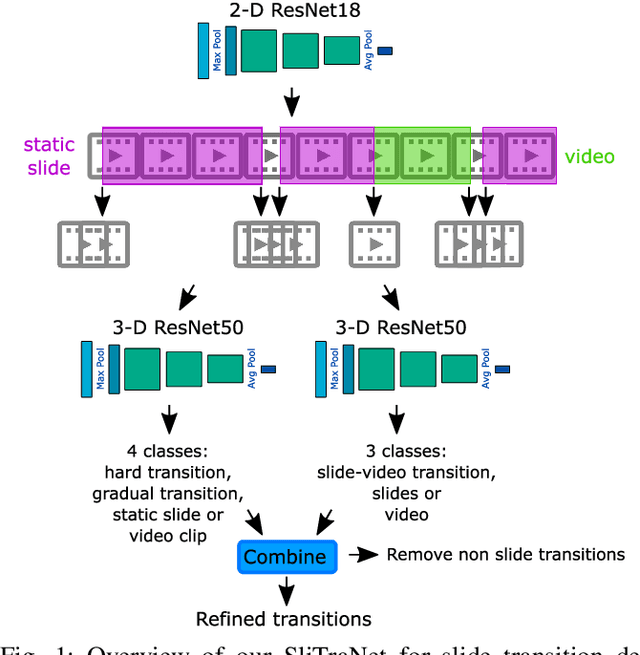
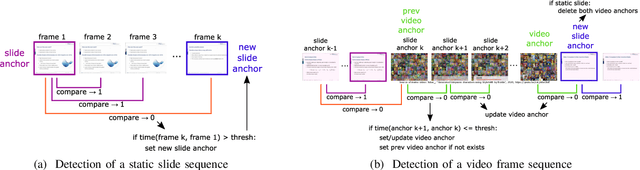
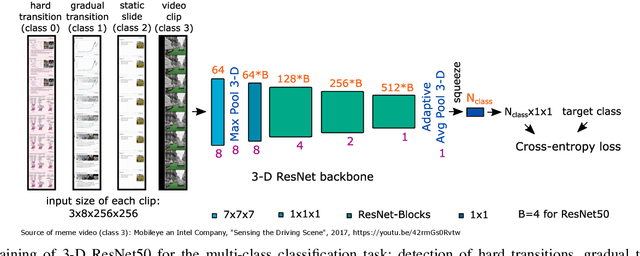

Abstract:With the increasing number of online learning material in the web, search for specific content in lecture videos can be time consuming. Therefore, automatic slide extraction from the lecture videos can be helpful to give a brief overview of the main content and to support the students in their studies. For this task, we propose a deep learning method to detect slide transitions in lectures videos. We first process each frame of the video by a heuristic-based approach using a 2-D convolutional neural network to predict transition candidates. Then, we increase the complexity by employing two 3-D convolutional neural networks to refine the transition candidates. Evaluation results demonstrate the effectiveness of our method in finding slide transitions.
A Keypoint Detection and Description Network Based on the Vessel Structure for Multi-Modal Retinal Image Registration
Jan 06, 2022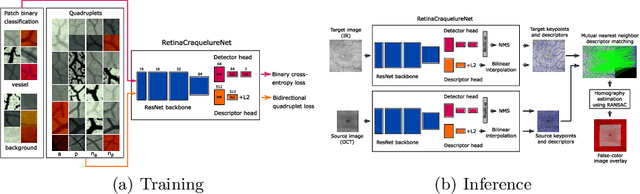

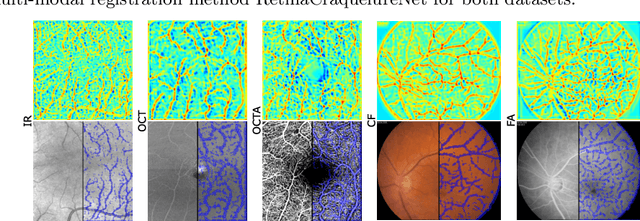
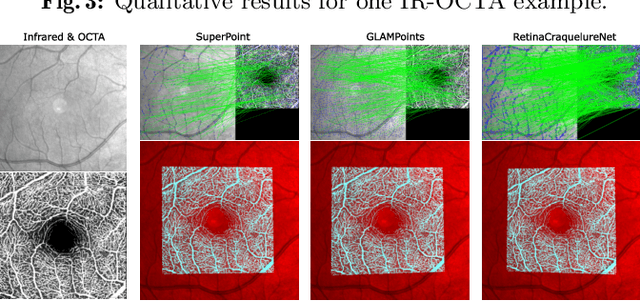
Abstract:Ophthalmological imaging utilizes different imaging systems, such as color fundus, infrared, fluorescein angiography, optical coherence tomography (OCT) or OCT angiography. Multiple images with different modalities or acquisition times are often analyzed for the diagnosis of retinal diseases. Automatically aligning the vessel structures in the images by means of multi-modal registration can support the ophthalmologists in their work. Our method uses a convolutional neural network to extract features of the vessel structure in multi-modal retinal images. We jointly train a keypoint detection and description network on small patches using a classification and a cross-modal descriptor loss function and apply the network to the full image size in the test phase. Our method demonstrates the best registration performance on our and a public multi-modal dataset in comparison to competing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge