Alexandre Andre
PRISM: A hierarchical multiscale approach for time series forecasting
Dec 31, 2025Abstract:Forecasting is critical in areas such as finance, biology, and healthcare. Despite the progress in the field, making accurate forecasts remains challenging because real-world time series contain both global trends, local fine-grained structure, and features on multiple scales in between. Here, we present a new forecasting method, PRISM (Partitioned Representation for Iterative Sequence Modeling), that addresses this challenge through a learnable tree-based partitioning of the signal. At the root of the tree, a global representation captures coarse trends in the signal, while recursive splits reveal increasingly localized views of the signal. At each level of the tree, data are projected onto a time-frequency basis (e.g., wavelets or exponential moving averages) to extract scale-specific features, which are then aggregated across the hierarchy. This design allows the model to jointly capture global structure and local dynamics of the signal, enabling accurate forecasting. Experiments across benchmark datasets show that our method outperforms state-of-the-art methods for forecasting. Overall, these results demonstrate that our hierarchical approach provides a lightweight and flexible framework for forecasting multivariate time series. The code is available at https://github.com/nerdslab/prism.
Neural Encoding and Decoding at Scale
Apr 14, 2025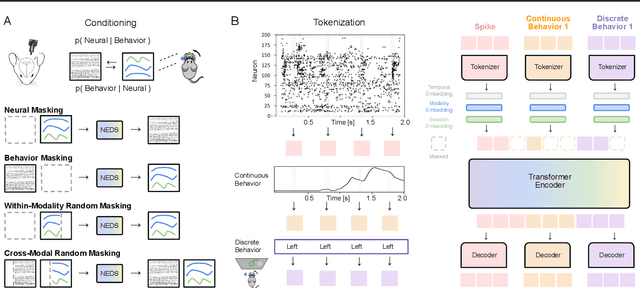
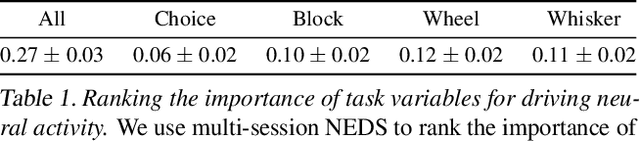
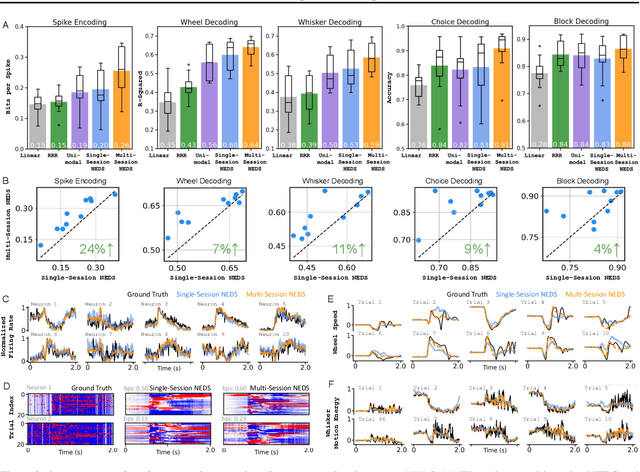
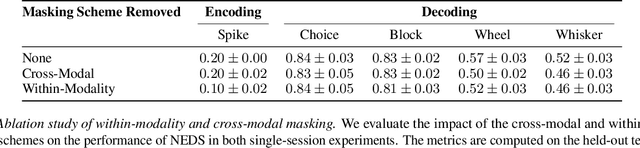
Abstract:Recent work has demonstrated that large-scale, multi-animal models are powerful tools for characterizing the relationship between neural activity and behavior. Current large-scale approaches, however, focus exclusively on either predicting neural activity from behavior (encoding) or predicting behavior from neural activity (decoding), limiting their ability to capture the bidirectional relationship between neural activity and behavior. To bridge this gap, we introduce a multimodal, multi-task model that enables simultaneous Neural Encoding and Decoding at Scale (NEDS). Central to our approach is a novel multi-task-masking strategy, which alternates between neural, behavioral, within-modality, and cross-modality masking. We pretrain our method on the International Brain Laboratory (IBL) repeated site dataset, which includes recordings from 83 animals performing the same visual decision-making task. In comparison to other large-scale models, we demonstrate that NEDS achieves state-of-the-art performance for both encoding and decoding when pretrained on multi-animal data and then fine-tuned on new animals. Surprisingly, NEDS's learned embeddings exhibit emergent properties: even without explicit training, they are highly predictive of the brain regions in each recording. Altogether, our approach is a step towards a foundation model of the brain that enables seamless translation between neural activity and behavior.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge