Alexander Matveev
Cross-Classification Clustering: An Efficient Multi-Object Tracking Technique for 3-D Instance Segmentation in Connectomics
Dec 04, 2018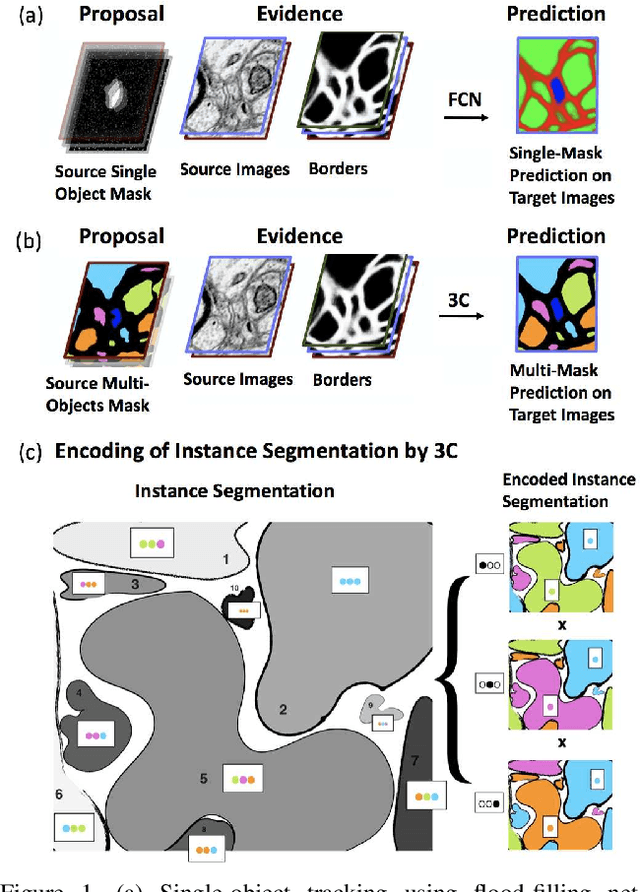
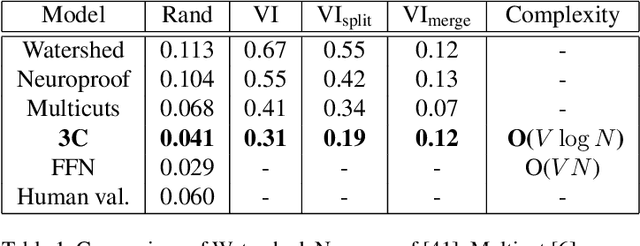
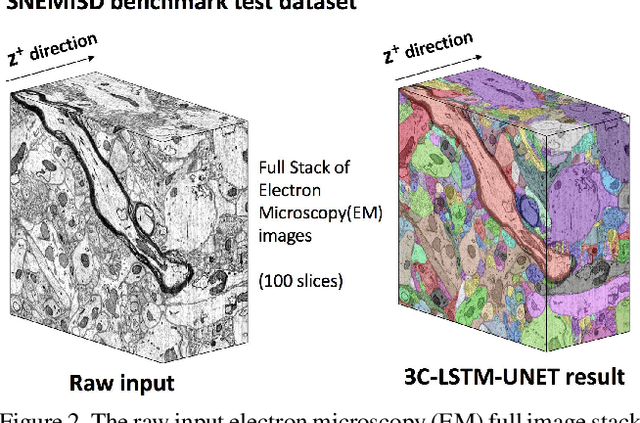
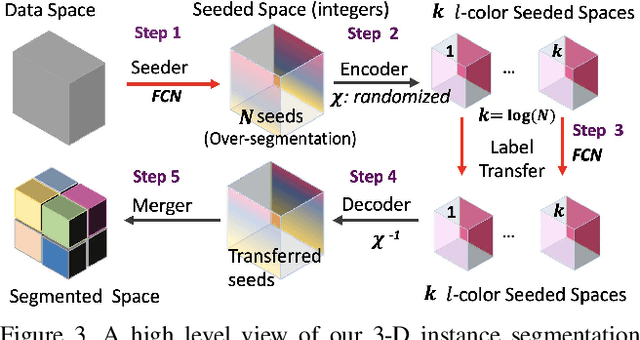
Abstract:Pixel-accurate tracking of objects is a key element in many computer vision applications, often solved by iterated individual object tracking or instance segmentation followed by object matching. Here we introduce cross-classification clustering (3C), a new technique that simultaneously tracks all objects in an image stack. The key idea in cross-classification is to efficiently turn a clustering problem into a classification problem by running a logarithmic number of independent classifications, letting the cross-labeling of these classifications uniquely classify each pixel to the object labels. We apply the 3C mechanism to achieve state-of-the-art accuracy in connectomics - nanoscale mapping of the brain from electron microscopy volumes. Our reconstruction system introduces an order of magnitude scalability improvement over the best current methods for neuronal reconstruction, and can be seamlessly integrated within existing single-object tracking methods like Google's flood-filling networks to improve their performance. This scalability is crucial for the real-world deployment of connectomics pipelines, as the best performing existing techniques require computing infrastructures that are beyond the reach of most labs. We believe 3C has valuable scalability implications in other domains that require pixel-accurate tracking of multiple objects in image stacks or video.
Deep Tensor Convolution on Multicores
Jun 11, 2017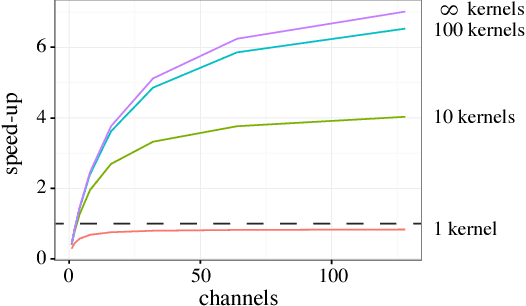
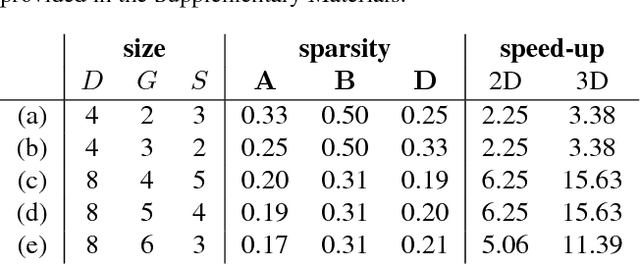
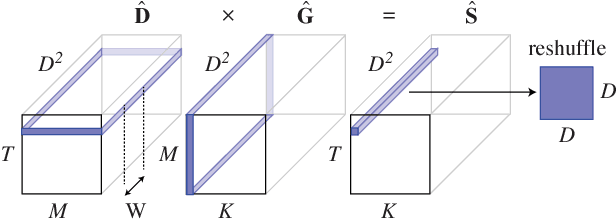
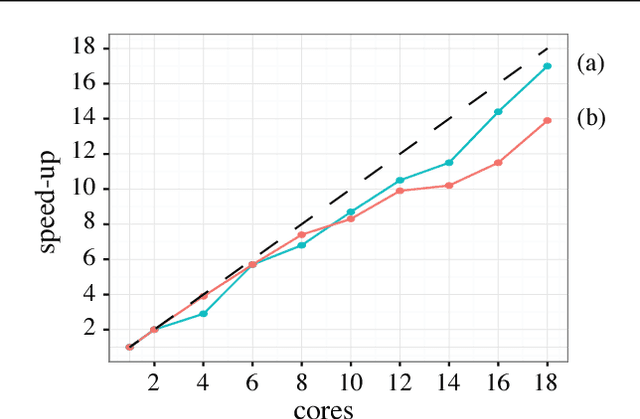
Abstract:Deep convolutional neural networks (ConvNets) of 3-dimensional kernels allow joint modeling of spatiotemporal features. These networks have improved performance of video and volumetric image analysis, but have been limited in size due to the low memory ceiling of GPU hardware. Existing CPU implementations overcome this constraint but are impractically slow. Here we extend and optimize the faster Winograd-class of convolutional algorithms to the $N$-dimensional case and specifically for CPU hardware. First, we remove the need to manually hand-craft algorithms by exploiting the relaxed constraints and cheap sparse access of CPU memory. Second, we maximize CPU utilization and multicore scalability by transforming data matrices to be cache-aware, integer multiples of AVX vector widths. Treating 2-dimensional ConvNets as a special (and the least beneficial) case of our approach, we demonstrate a 5 to 25-fold improvement in throughput compared to previous state-of-the-art.
Toward Streaming Synapse Detection with Compositional ConvNets
Feb 23, 2017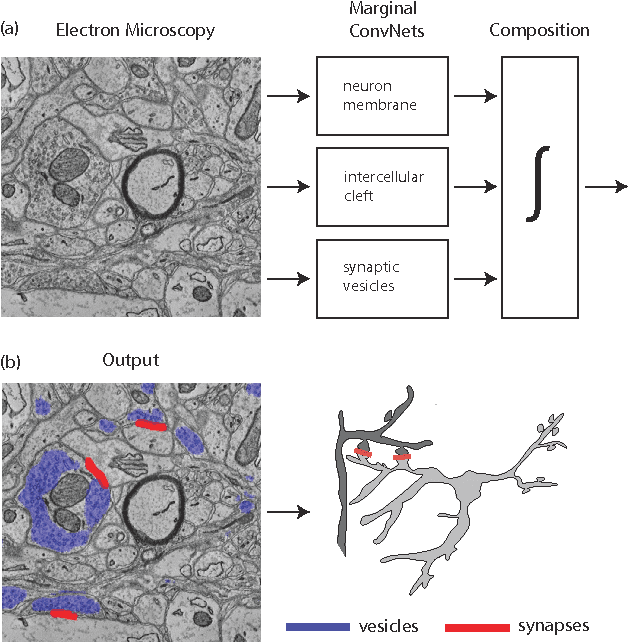
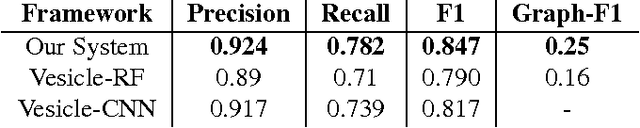
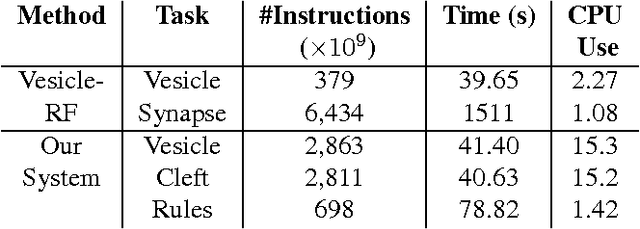
Abstract:Connectomics is an emerging field in neuroscience that aims to reconstruct the 3-dimensional morphology of neurons from electron microscopy (EM) images. Recent studies have successfully demonstrated the use of convolutional neural networks (ConvNets) for segmenting cell membranes to individuate neurons. However, there has been comparatively little success in high-throughput identification of the intercellular synaptic connections required for deriving connectivity graphs. In this study, we take a compositional approach to segmenting synapses, modeling them explicitly as an intercellular cleft co-located with an asymmetric vesicle density along a cell membrane. Instead of requiring a deep network to learn all natural combinations of this compositionality, we train lighter networks to model the simpler marginal distributions of membranes, clefts and vesicles from just 100 electron microscopy samples. These feature maps are then combined with simple rules-based heuristics derived from prior biological knowledge. Our approach to synapse detection is both more accurate than previous state-of-the-art (7% higher recall and 5% higher F1-score) and yields a 20-fold speed-up compared to the previous fastest implementations. We demonstrate by reconstructing the first complete, directed connectome from the largest available anisotropic microscopy dataset (245 GB) of mouse somatosensory cortex (S1) in just 9.7 hours on a single shared-memory CPU system. We believe that this work marks an important step toward the goal of a microscope-pace streaming connectomics pipeline.
A Multi-Pass Approach to Large-Scale Connectomics
Dec 07, 2016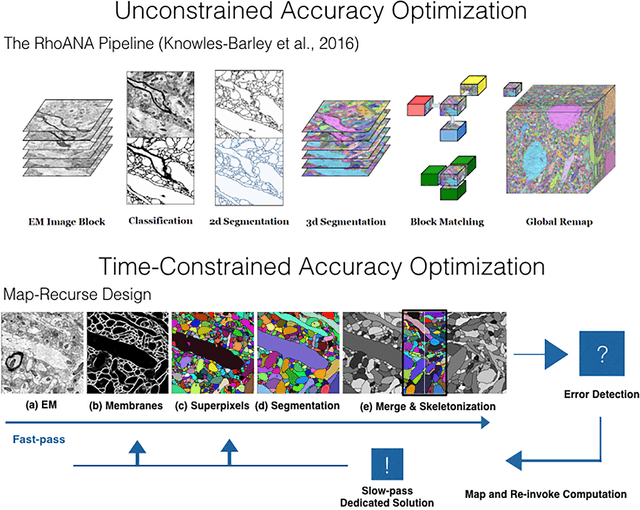
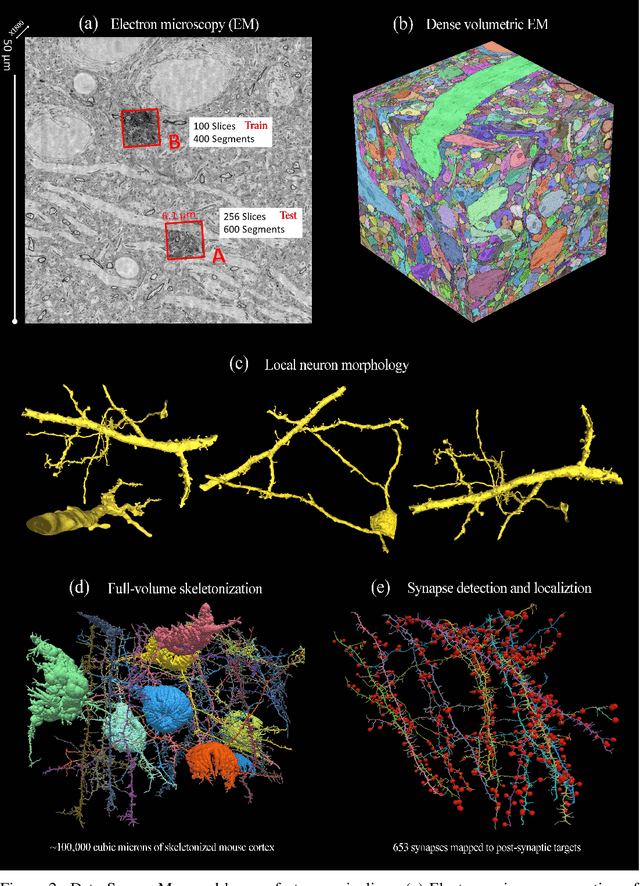
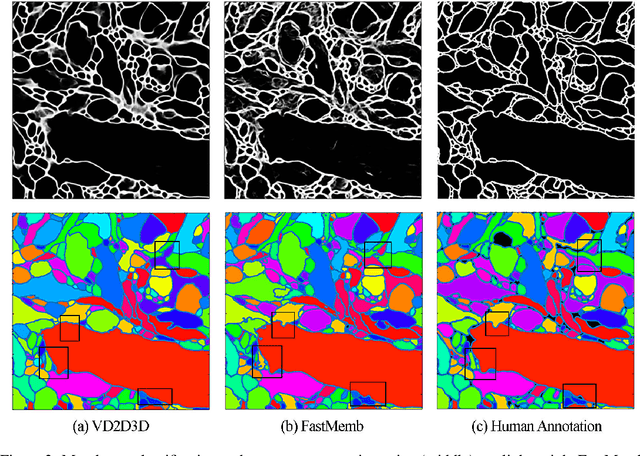
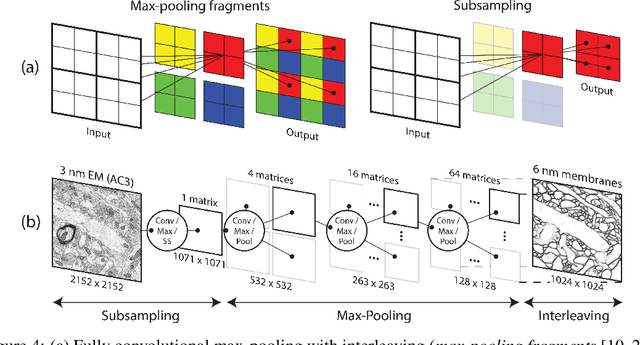
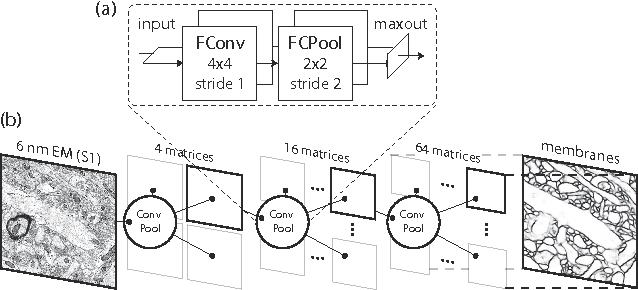
Abstract:The field of connectomics faces unprecedented "big data" challenges. To reconstruct neuronal connectivity, automated pixel-level segmentation is required for petabytes of streaming electron microscopy data. Existing algorithms provide relatively good accuracy but are unacceptably slow, and would require years to extract connectivity graphs from even a single cubic millimeter of neural tissue. Here we present a viable real-time solution, a multi-pass pipeline optimized for shared-memory multicore systems, capable of processing data at near the terabyte-per-hour pace of multi-beam electron microscopes. The pipeline makes an initial fast-pass over the data, and then makes a second slow-pass to iteratively correct errors in the output of the fast-pass. We demonstrate the accuracy of a sparse slow-pass reconstruction algorithm and suggest new methods for detecting morphological errors. Our fast-pass approach provided many algorithmic challenges, including the design and implementation of novel shallow convolutional neural nets and the parallelization of watershed and object-merging techniques. We use it to reconstruct, from image stack to skeletons, the full dataset of Kasthuri et al. (463 GB capturing 120,000 cubic microns) in a matter of hours on a single multicore machine rather than the weeks it has taken in the past on much larger distributed systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge