Achille Salaün
Sample Selection Bias in Machine Learning for Healthcare
May 13, 2024



Abstract:While machine learning algorithms hold promise for personalised medicine, their clinical adoption remains limited. One critical factor contributing to this restraint is sample selection bias (SSB) which refers to the study population being less representative of the target population, leading to biased and potentially harmful decisions. Despite being well-known in the literature, SSB remains scarcely studied in machine learning for healthcare. Moreover, the existing techniques try to correct the bias by balancing distributions between the study and the target populations, which may result in a loss of predictive performance. To address these problems, our study illustrates the potential risks associated with SSB by examining SSB's impact on the performance of machine learning algorithms. Most importantly, we propose a new research direction for addressing SSB, based on the target population identification rather than the bias correction. Specifically, we propose two independent networks (T-Net) and a multitasking network (MT-Net) for addressing SSB, where one network/task identifies the target subpopulation which is representative of the study population and the second makes predictions for the identified subpopulation. Our empirical results with synthetic and semi-synthetic datasets highlight that SSB can lead to a large drop in the performance of an algorithm for the target population as compared with the study population, as well as a substantial difference in the performance for the target subpopulations that are representative of the selected and the non-selected patients from the study population. Furthermore, our proposed techniques demonstrate robustness across various settings, including different dataset sizes, event rates, and selection rates, outperforming the existing bias correction techniques.
Expressivity of Hidden Markov Chains vs. Recurrent Neural Networks from a system theoretic viewpoint
Aug 17, 2022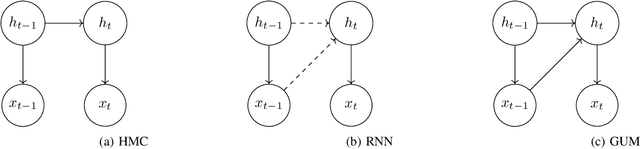

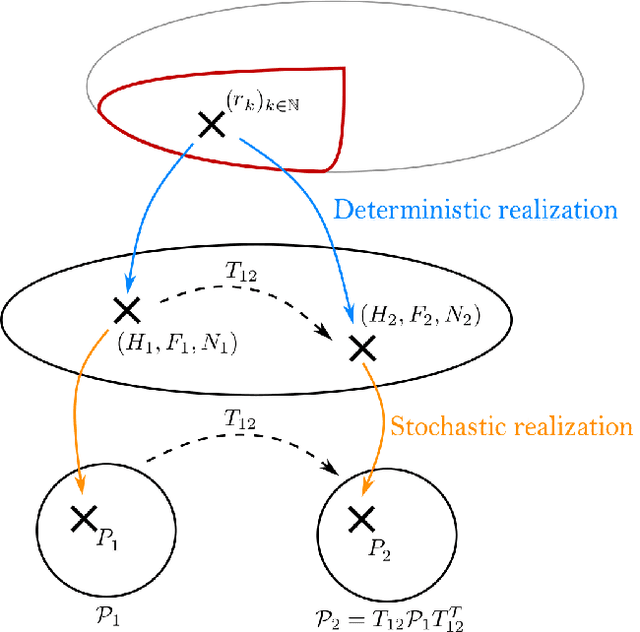
Abstract:Hidden Markov Chains (HMC) and Recurrent Neural Networks (RNN) are two well known tools for predicting time series. Even though these solutions were developed independently in distinct communities, they share some similarities when considered as probabilistic structures. So in this paper we first consider HMC and RNN as generative models, and we embed both structures in a common generative unified model (GUM). We next address a comparative study of the expressivity of these models. To that end we assume that the models are furthermore linear and Gaussian. The probability distributions produced by these models are characterized by structured covariance series, and as a consequence expressivity reduces to comparing sets of structured covariance series, which enables us to call for stochastic realization theory (SRT). We finally provide conditions under which a given covariance series can be realized by a GUM, an HMC or an RNN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge