Abdullah Nazib
Enhancing Semantic Segmentation with Adaptive Focal Loss: A Novel Approach
Jul 13, 2024Abstract:Deep learning has achieved outstanding accuracy in medical image segmentation, particularly for objects like organs or tumors with smooth boundaries or large sizes. Whereas, it encounters significant difficulties with objects that have zigzag boundaries or are small in size, leading to a notable decrease in segmentation effectiveness. In this context, using a loss function that incorporates smoothness and volume information into a model's predictions offers a promising solution to these shortcomings. In this work, we introduce an Adaptive Focal Loss (A-FL) function designed to mitigate class imbalance by down-weighting the loss for easy examples that results in up-weighting the loss for hard examples and giving greater emphasis to challenging examples, such as small and irregularly shaped objects. The proposed A-FL involves dynamically adjusting a focusing parameter based on an object's surface smoothness, size information, and adjusting the class balancing parameter based on the ratio of targeted area to total area in an image. We evaluated the performance of the A-FL using ResNet50-encoded U-Net architecture on the Picai 2022 and BraTS 2018 datasets. On the Picai 2022 dataset, the A-FL achieved an Intersection over Union (IoU) of 0.696 and a Dice Similarity Coefficient (DSC) of 0.769, outperforming the regular Focal Loss (FL) by 5.5% and 5.4% respectively. It also surpassed the best baseline Dice-Focal by 2.0% and 1.2%. On the BraTS 2018 dataset, A-FL achieved an IoU of 0.883 and a DSC of 0.931. The comparative studies show that the proposed A-FL function surpasses conventional methods, including Dice Loss, Focal Loss, and their hybrid variants, in IoU, DSC, Sensitivity, and Specificity metrics. This work highlights A-FL's potential to improve deep learning models for segmenting clinically significant regions in medical images, leading to more precise and reliable diagnostic tools.
Uncertainty Driven Bottleneck Attention U-net for OAR Segmentation
Mar 19, 2023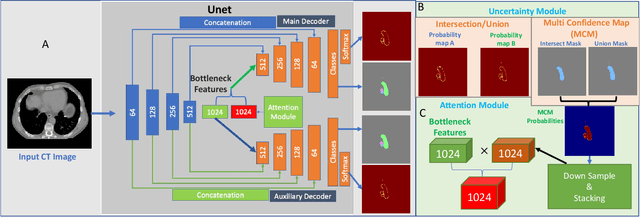
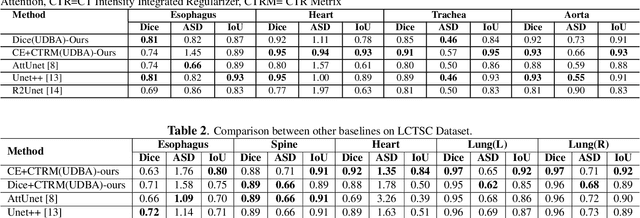
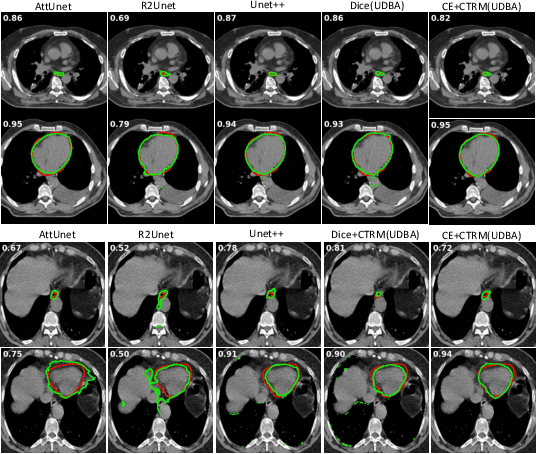
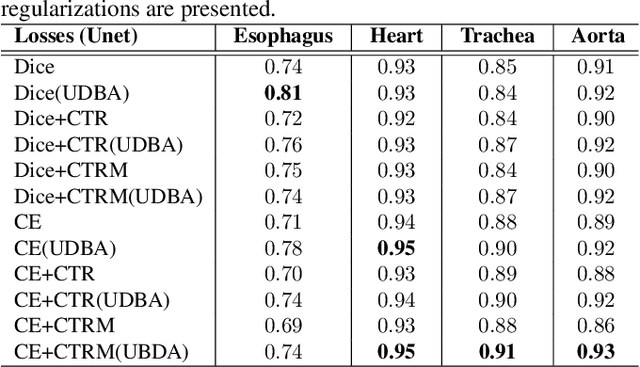
Abstract:Organ at risk (OAR) segmentation in computed tomography (CT) imagery is a difficult task for automated segmentation methods and can be crucial for downstream radiation treatment planning. U-net has become a de-facto standard for medical image segmentation and is frequently used as a common baseline in medical image segmentation tasks. In this paper, we develop a multiple decoder U-net architecture where a noisy auxiliary decoder is used to generate noisy segmentation. The segmentation from the main branch and the noisy segmentation from the auxiliary branch are used together to estimate the attention. Our contribution is the development of a new attention module which derives the attention from the softmax probabilities of two decoder branches. The union and intersection of two segmentation masks from two branches carry the information where both decoders agree and disagree. The softmax probabilities from regions of agreement and disagreement are the indicators of low and high uncertainty. Thus, the probabilities of those selected regions are used as attention in the bottleneck layer of the encoder and passes only through the main decoder for segmentation. For accurate contour segmentation, we also developed a CT intensity integrated regularization loss. We tested our model on two publicly available OAR challenge datasets, Segthor and LCTSC respectively. We trained 12 models on each dataset with and without the proposed attention model and regularization loss to check the effectiveness of the attention module and the regularization loss. The experiments demonstrate a clear accuracy improvement (2\% to 5\% Dice) on both datasets. Code for the experiments will be made available upon the acceptance for publication.
A Multiple Decoder CNN for Inverse Consistent 3D Image Registration
Feb 15, 2020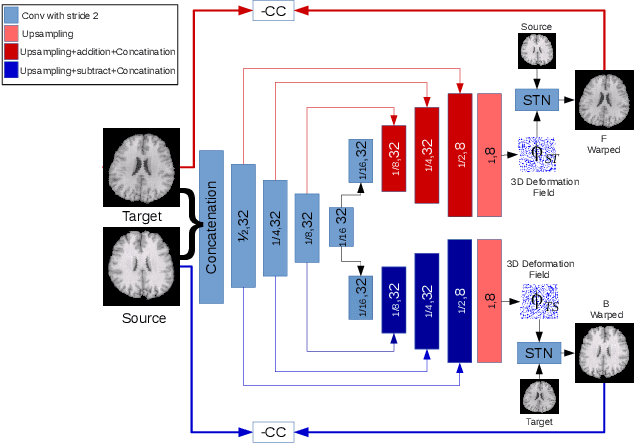

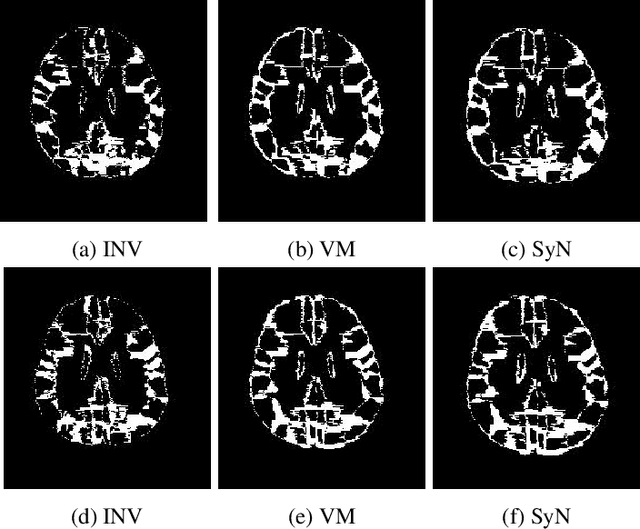
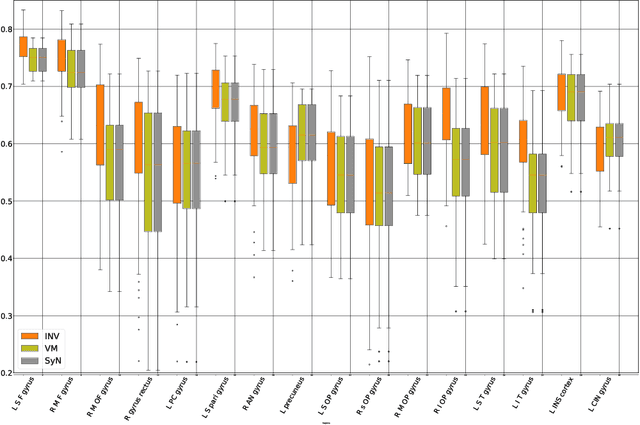
Abstract:The recent application of deep learning technologies in medical image registration has exponentially decreased the registration time and gradually increased registration accuracy when compared to their traditional counterparts. Most of the learning-based registration approaches considers this task as a one directional problem. As a result, only correspondence from the moving image to the target image is considered. However, in some medical procedures bidirectional registration is required to be performed. Unlike other learning-based registration, we propose a registration framework with inverse consistency. The proposed method simultaneously learns forward transformation and backward transformation in an unsupervised manner. We perform training and testing of the method on the publicly available LPBA40 MRI dataset and demonstrate strong performance than baseline registration methods.
Dense Deformation Network for High Resolution Tissue Cleared Image Registration
Jun 13, 2019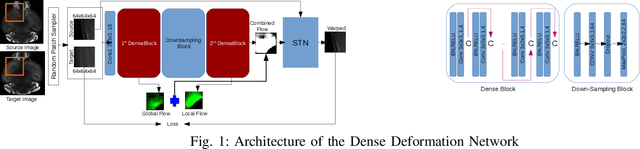
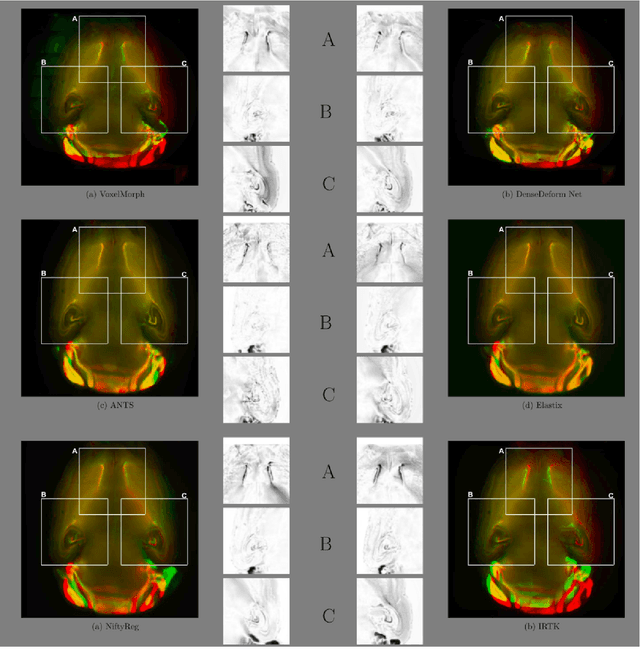
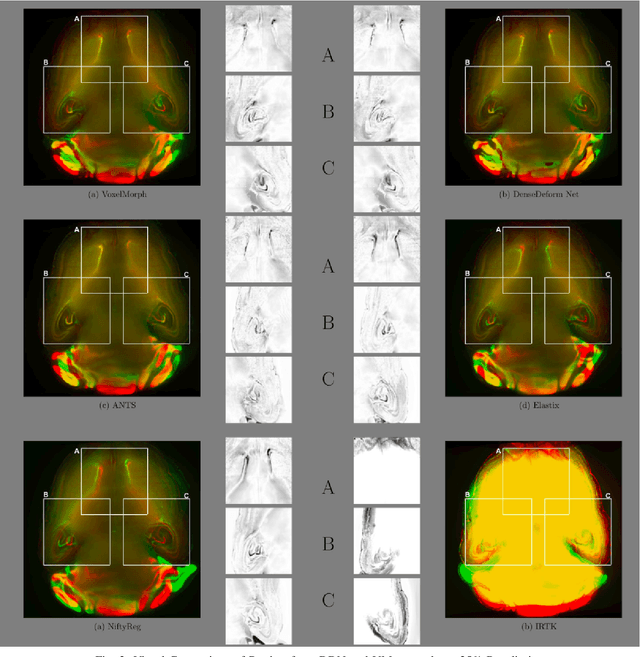
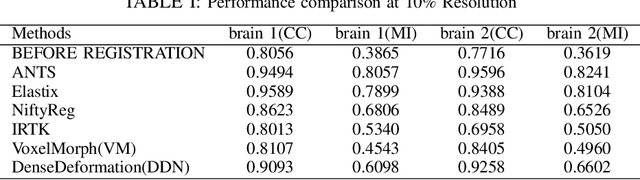
Abstract:The recent application of Deep Learning in various areas of medical image analysis has brought excellent performance gain. The application of deep learning technologies in medical image registration successfully outperformed traditional optimization based registration algorithms both in registration time and accuracy. In this paper, we present a densely connected convolutional architecture for deformable image registration. The training of the network is unsupervised and does not require ground-truth deformation or any synthetic deformation as a label. The proposed architecture is trained and tested on two different version of tissue cleared data, 10\% and 25\% resolution of high resolution dataset respectively and demonstrated comparable registration performance with the state-of-the-art ANTS registration method. The proposed method is also compared with the deep-learning based Voxelmorph registration method. Due to the memory limitation, original voxelmorph can work at most 15\% resolution of Tissue cleared data. For rigorous experimental comparison we developed a patch-based version of Voxelmorph network, and trained it on 10\% and 25\% resolution. In both resolution, proposed DenseDeformation network outperformed Voxelmorph in registration accuracy.
A Comparative Analysis of Registration Tools: Traditional vs Deep Learning Approach on High Resolution Tissue Cleared Data
Oct 19, 2018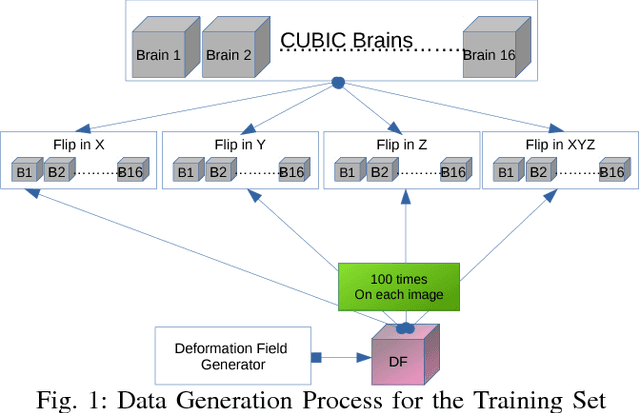
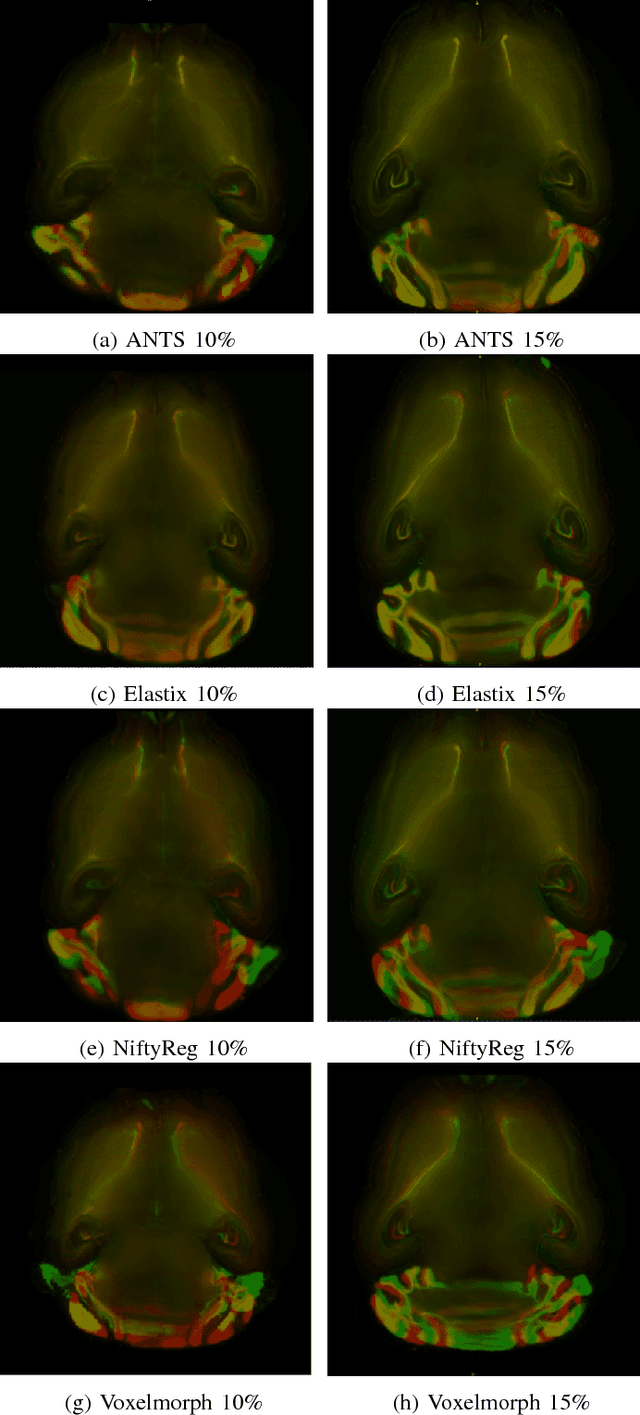
Abstract:Image registration plays an important role in comparing images. It is particularly important in analyzing medical images like CT, MRI, PET, etc. to quantify different biological samples, to monitor disease progression and to fuse different modalities to support better diagnosis. The recent emergence of tissue clearing protocols enable us to take images at cellular level resolution. Image registration tools developed for other modalities are currently unable to manage images of such extreme high resolution. The recent popularity of deep learning based methods in the computer vision community justifies a rigorous investigation of deep-learning based methods on tissue cleared images along with their traditional counterparts. In this paper, we investigate and compare the performance of a deep learning based registration method with traditional optimization based methods on samples from tissue-clearing methods. From the comparative results it is found that a deep-learning based method outperforms all traditional registration tools in terms of registration time and has achieved promising registration accuracy.
Performance of Image Registration Tools on High-Resolution 3D Brain Images
Jul 13, 2018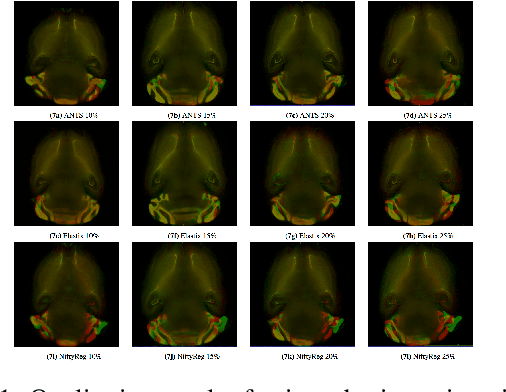
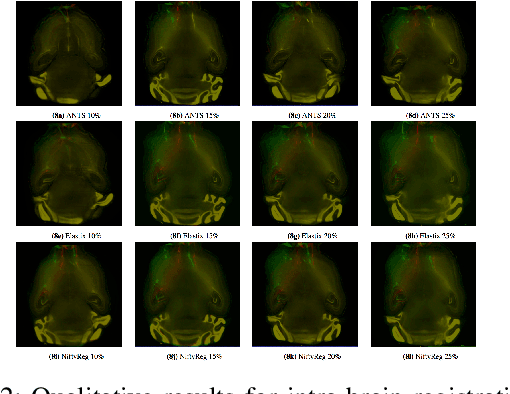
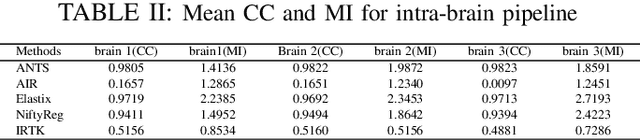
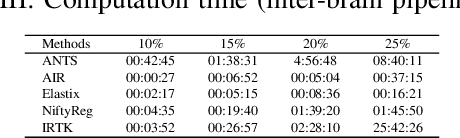
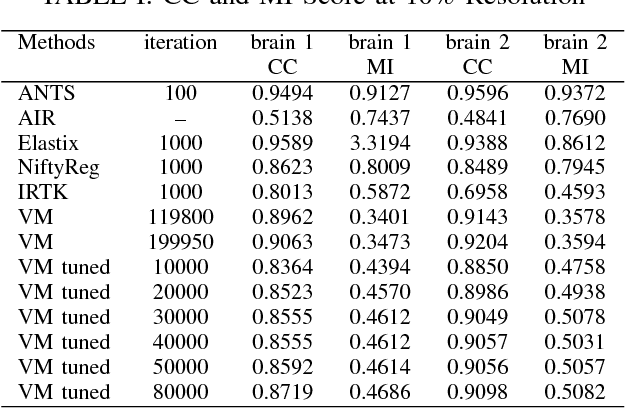
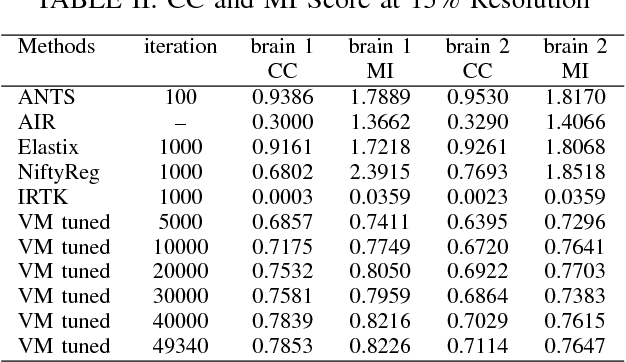
Abstract:Recent progress in tissue clearing has allowed for the imaging of entire organs at single-cell resolution. These methods produce very large 3D images (several gigabytes for a whole mouse brain). A necessary step in analysing these images is registration across samples. Existing methods of registration were developed for lower resolution image modalities (e.g. MRI) and it is unclear whether their performance and accuracy is satisfactory at this larger scale. In this study, we used data from different mouse brains cleared with the CUBIC protocol to evaluate five freely available image registration tools. We used several performance metrics to assess accuracy, and completion time as a measure of efficiency. The results of this evaluation suggest that the ANTS registration tool provides the best registration accuracy while Elastix has the highest computational efficiency among the methods with an acceptable accuracy. The results also highlight the need to develop new registration methods optimised for these high-resolution 3D images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge