Dimitri Perrin
An Adversarial Approach to Register Extreme Resolution Tissue Cleared 3D Brain Images
Jan 31, 2025Abstract:We developed a generative patch based 3D image registration model that can register very high resolution images obtained from a biochemical process name tissue clearing. Tissue clearing process removes lipids and fats from the tissue and make the tissue transparent. When cleared tissues are imaged with Light-sheet fluorescent microscopy, the resulting images give a clear window to the cellular activities and dynamics inside the tissue.Thus the images obtained are very rich with cellular information and hence their resolution is extremely high (eg .2560x2160x676). Analyzing images with such high resolution is a difficult task for any image analysis pipeline.Image registration is a common step in image analysis pipeline when comparison between images are required. Traditional image registration methods fail to register images with such extant. In this paper we addressed this very high resolution image registration issue by proposing a patch-based generative network named InvGAN. Our proposed network can register very high resolution tissue cleared images. The tissue cleared dataset used in this paper are obtained from a tissue clearing protocol named CUBIC. We compared our method both with traditional and deep-learning based registration methods.Two different versions of CUBIC dataset are used, representing two different resolutions 25% and 100% respectively. Experiments on two different resolutions clearly show the impact of resolution on the registration quality. At 25% resolution, our method achieves comparable registration accuracy with very short time (7 minutes approximately). At 100% resolution, most of the traditional registration methods fail except Elastix registration tool.Elastix takes 28 hours to register where proposed InvGAN takes only 10 minutes.
Piecewise Deterministic Markov Processes for Bayesian Neural Networks
Feb 17, 2023Abstract:Inference on modern Bayesian Neural Networks (BNNs) often relies on a variational inference treatment, imposing violated assumptions of independence and the form of the posterior. Traditional MCMC approaches avoid these assumptions at the cost of increased computation due to its incompatibility to subsampling of the likelihood. New Piecewise Deterministic Markov Process (PDMP) samplers permit subsampling, though introduce a model specific inhomogenous Poisson Process (IPPs) which is difficult to sample from. This work introduces a new generic and adaptive thinning scheme for sampling from these IPPs, and demonstrates how this approach can accelerate the application of PDMPs for inference in BNNs. Experimentation illustrates how inference with these methods is computationally feasible, can improve predictive accuracy, MCMC mixing performance, and provide informative uncertainty measurements when compared against other approximate inference schemes.
A Multiple Decoder CNN for Inverse Consistent 3D Image Registration
Feb 15, 2020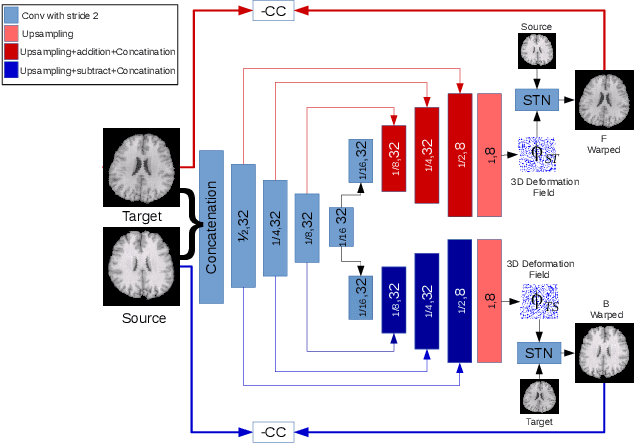

Abstract:The recent application of deep learning technologies in medical image registration has exponentially decreased the registration time and gradually increased registration accuracy when compared to their traditional counterparts. Most of the learning-based registration approaches considers this task as a one directional problem. As a result, only correspondence from the moving image to the target image is considered. However, in some medical procedures bidirectional registration is required to be performed. Unlike other learning-based registration, we propose a registration framework with inverse consistency. The proposed method simultaneously learns forward transformation and backward transformation in an unsupervised manner. We perform training and testing of the method on the publicly available LPBA40 MRI dataset and demonstrate strong performance than baseline registration methods.
Dense Deformation Network for High Resolution Tissue Cleared Image Registration
Jun 13, 2019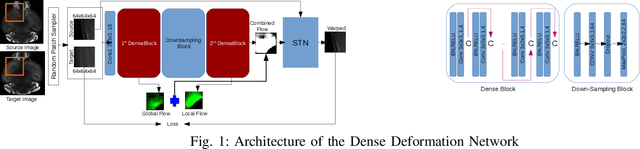
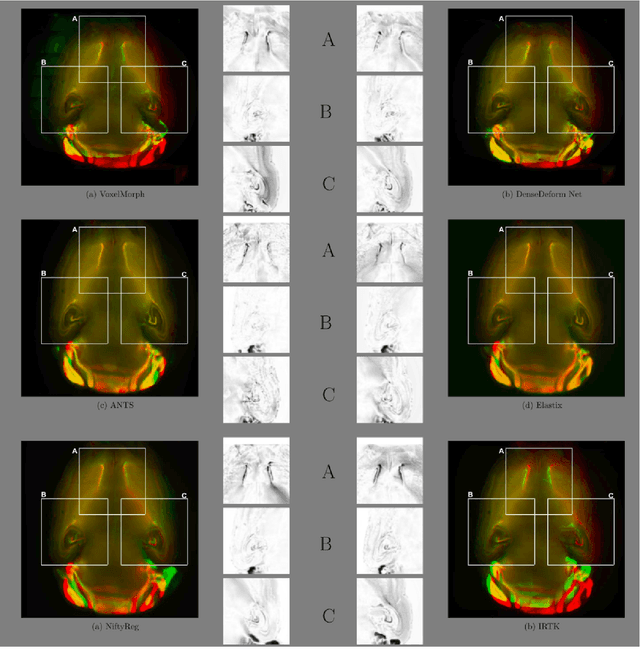
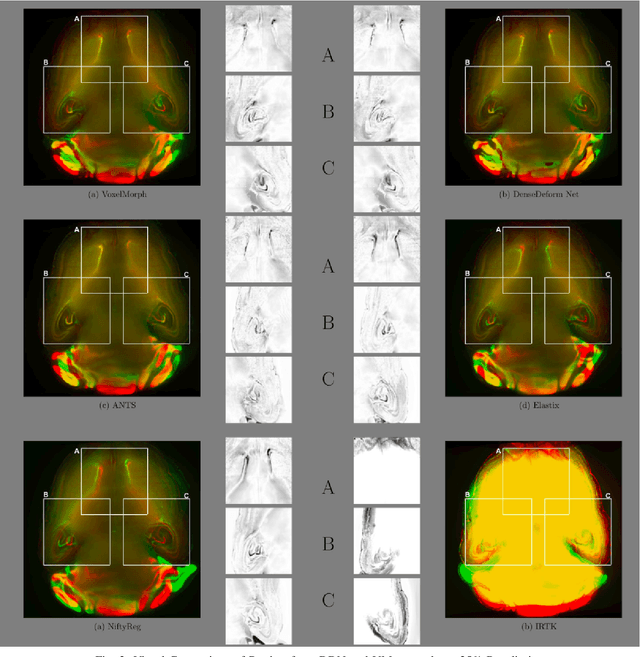
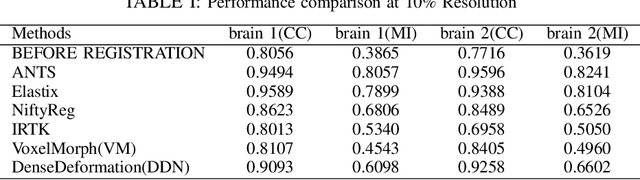
Abstract:The recent application of Deep Learning in various areas of medical image analysis has brought excellent performance gain. The application of deep learning technologies in medical image registration successfully outperformed traditional optimization based registration algorithms both in registration time and accuracy. In this paper, we present a densely connected convolutional architecture for deformable image registration. The training of the network is unsupervised and does not require ground-truth deformation or any synthetic deformation as a label. The proposed architecture is trained and tested on two different version of tissue cleared data, 10\% and 25\% resolution of high resolution dataset respectively and demonstrated comparable registration performance with the state-of-the-art ANTS registration method. The proposed method is also compared with the deep-learning based Voxelmorph registration method. Due to the memory limitation, original voxelmorph can work at most 15\% resolution of Tissue cleared data. For rigorous experimental comparison we developed a patch-based version of Voxelmorph network, and trained it on 10\% and 25\% resolution. In both resolution, proposed DenseDeformation network outperformed Voxelmorph in registration accuracy.
A Comparative Analysis of Registration Tools: Traditional vs Deep Learning Approach on High Resolution Tissue Cleared Data
Oct 19, 2018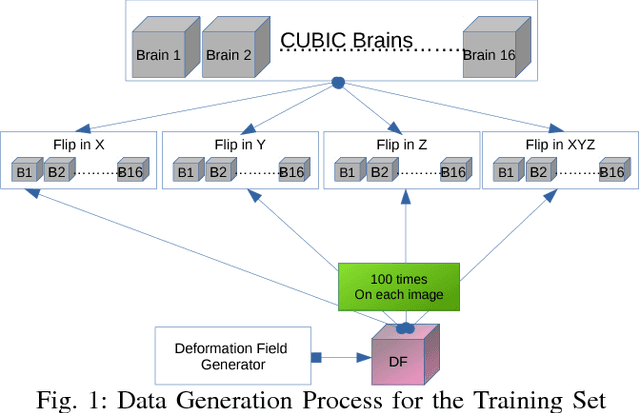
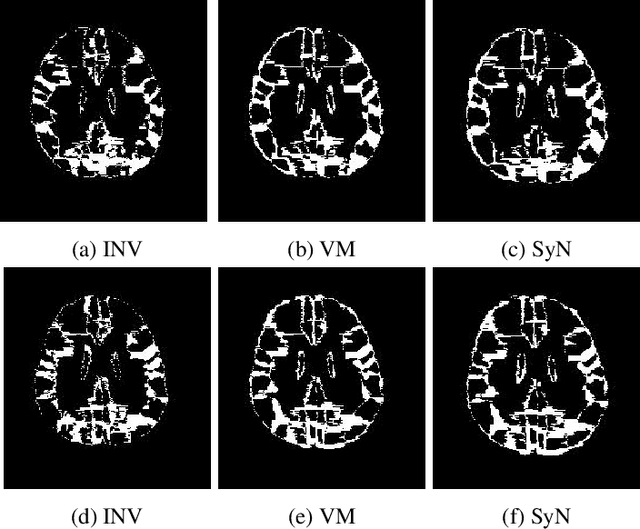
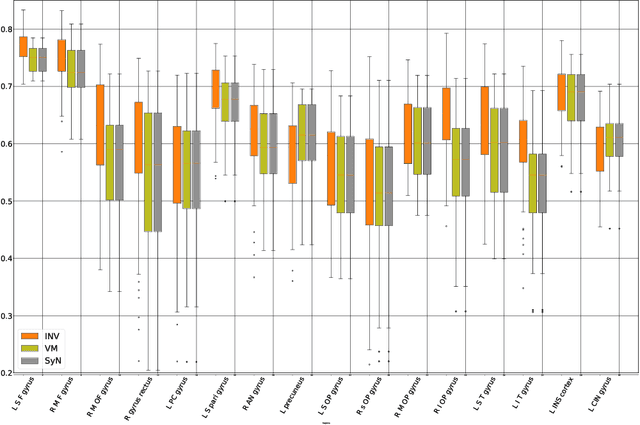
Abstract:Image registration plays an important role in comparing images. It is particularly important in analyzing medical images like CT, MRI, PET, etc. to quantify different biological samples, to monitor disease progression and to fuse different modalities to support better diagnosis. The recent emergence of tissue clearing protocols enable us to take images at cellular level resolution. Image registration tools developed for other modalities are currently unable to manage images of such extreme high resolution. The recent popularity of deep learning based methods in the computer vision community justifies a rigorous investigation of deep-learning based methods on tissue cleared images along with their traditional counterparts. In this paper, we investigate and compare the performance of a deep learning based registration method with traditional optimization based methods on samples from tissue-clearing methods. From the comparative results it is found that a deep-learning based method outperforms all traditional registration tools in terms of registration time and has achieved promising registration accuracy.
Performance of Image Registration Tools on High-Resolution 3D Brain Images
Jul 13, 2018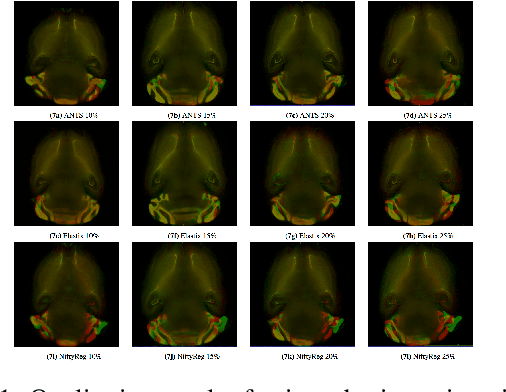
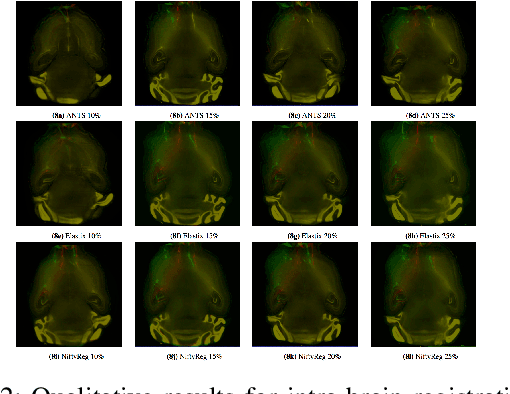
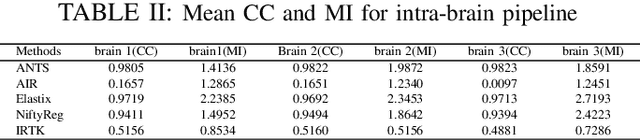
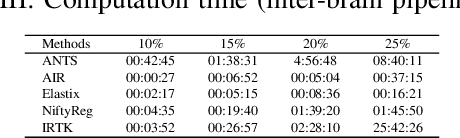
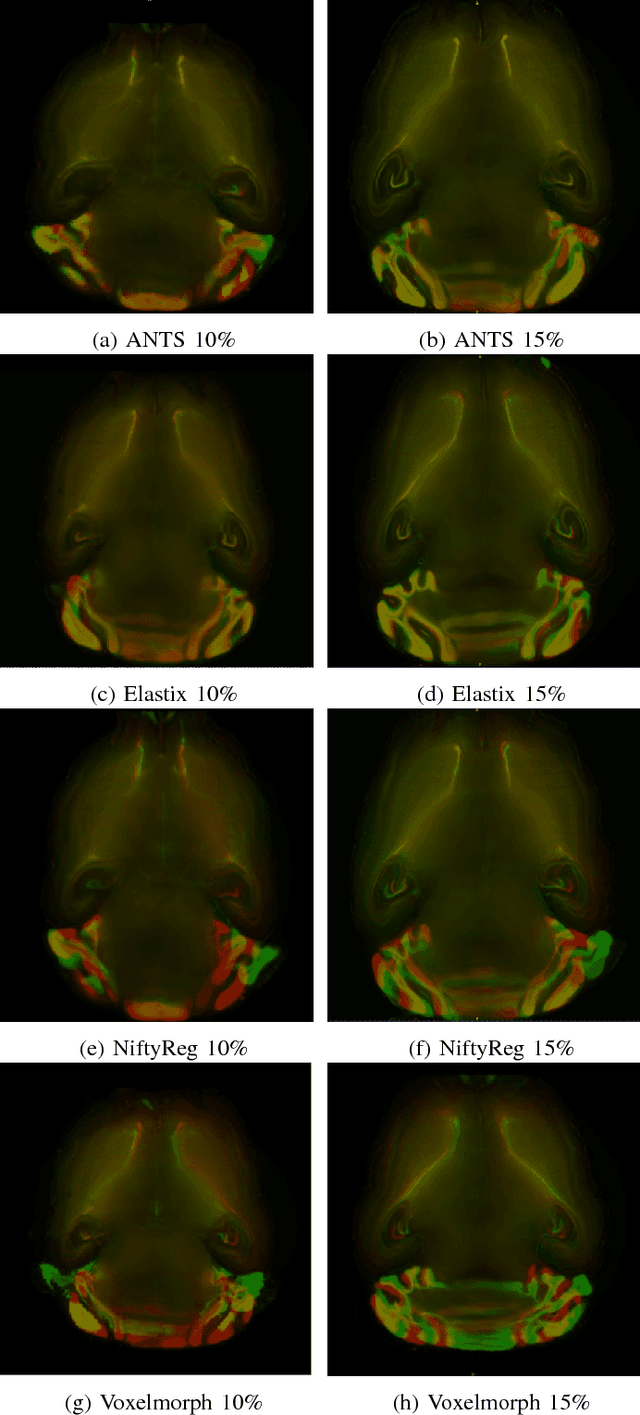
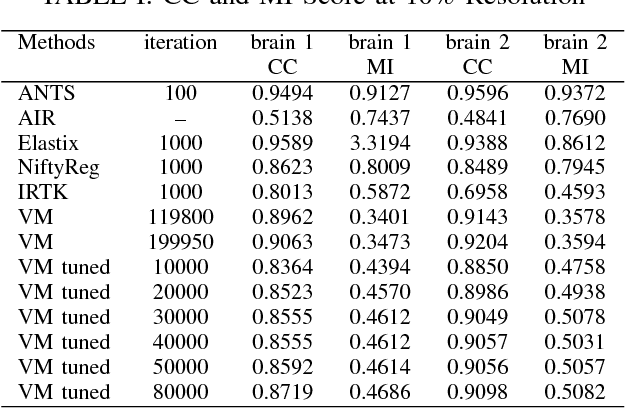
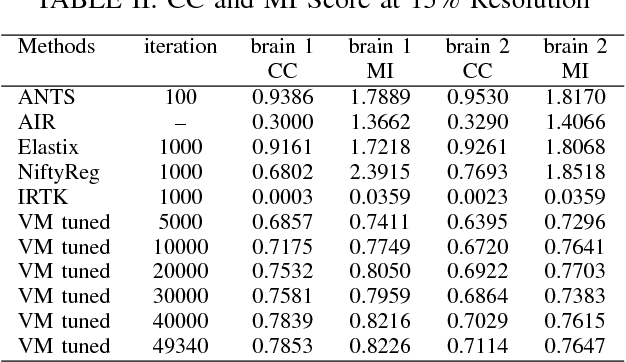
Abstract:Recent progress in tissue clearing has allowed for the imaging of entire organs at single-cell resolution. These methods produce very large 3D images (several gigabytes for a whole mouse brain). A necessary step in analysing these images is registration across samples. Existing methods of registration were developed for lower resolution image modalities (e.g. MRI) and it is unclear whether their performance and accuracy is satisfactory at this larger scale. In this study, we used data from different mouse brains cleared with the CUBIC protocol to evaluate five freely available image registration tools. We used several performance metrics to assess accuracy, and completion time as a measure of efficiency. The results of this evaluation suggest that the ANTS registration tool provides the best registration accuracy while Elastix has the highest computational efficiency among the methods with an acceptable accuracy. The results also highlight the need to develop new registration methods optimised for these high-resolution 3D images.
The use of neural networks in the analysis of sleep stages and the diagnosis of narcolepsy
Oct 05, 2017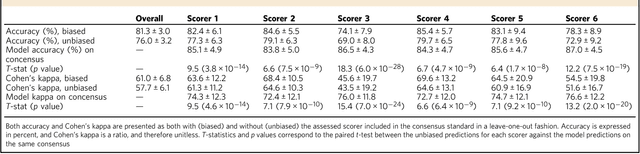
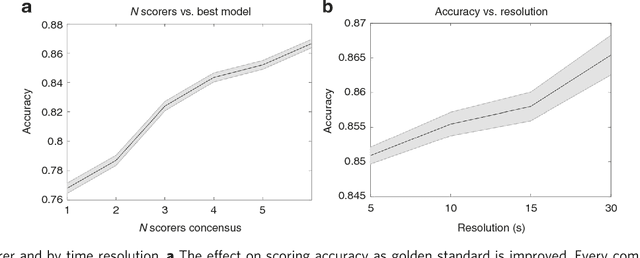

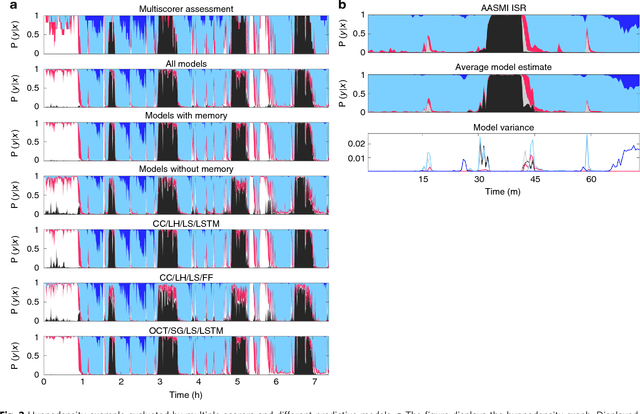
Abstract:We used neural networks in ~3,000 sleep recordings from over 10 locations to automate sleep stage scoring, producing a probability distribution called an hypnodensity graph. Accuracy was validated in 70 subjects scored by six technicians (gold standard). Our best model performed better than any individual scorer, reaching an accuracy of 0.87 (and 0.95 when predictions are weighed by scorer agreement). It also scores sleep stages down to 5-second instead of the conventional 30-second scoring-epochs. Accuracy did not vary by sleep disorder except for narcolepsy, suggesting scoring difficulties by machine and/or humans. A narcolepsy biomarker was extracted and validated in 105 type-1 narcoleptics versus 331 controls producing a specificity of 0.96 and a sensitivity of 0.91. Similar performances were obtained against a high pretest probability sample of type-2 narcolepsy and idiopathic hypersomnia patients. Addition of HLA-DQB1*06:02 increased specificity to 0.99. Our method streamlines scoring and diagnoses narcolepsy accurately.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge