Revealing Fine Structures of the Retinal Receptive Field by Deep Learning Networks
Paper and Code
Nov 06, 2018
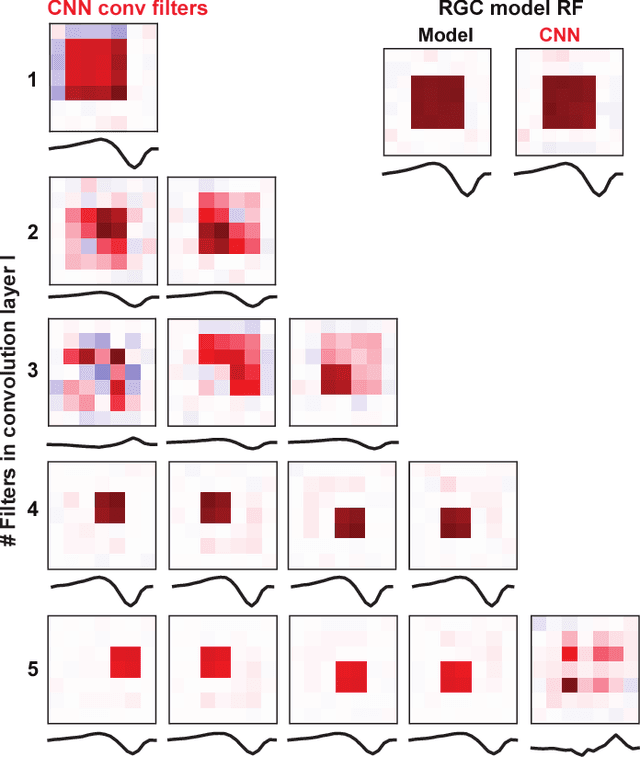
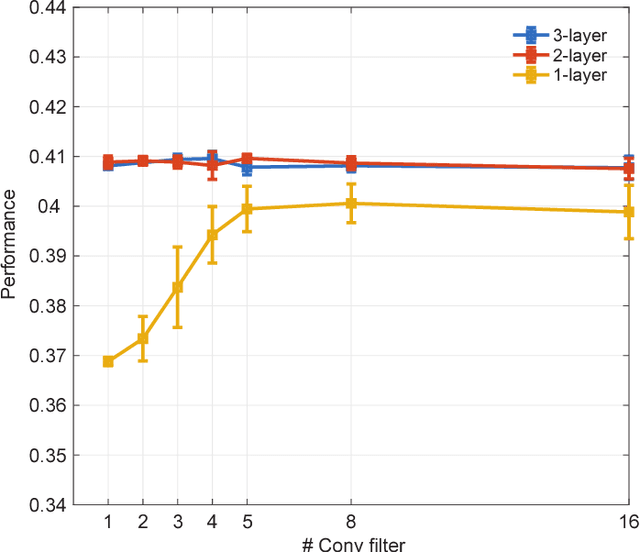
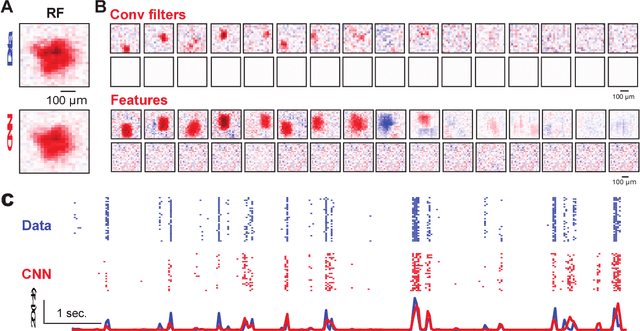
Deep convolutional neural networks (CNNs) have demonstrated impressive performance on many visual tasks. Recently, they became useful models for the visual system in neuroscience. However, it is still not clear what are learned by CNNs in terms of neuronal circuits. When a deep CNN with many layers is used for the visual system, it is not easy to compare the structure components of CNN with possible neuroscience underpinnings due to highly complex circuits from the retina to higher visual cortex. Here we address this issue by focusing on single retinal ganglion cells with biophysical models and recording data from animals. By training CNNs with white noise images to predict neuronal responses, we found that fine structures of the retinal receptive field can be revealed. Specifically, convolutional filters learned are resembling biological components of the retinal circuit. This suggests that a CNN learning from one single retinal cell reveals a minimal neural network carried out in this cell. Furthermore, when CNNs learned from different cells are transferred between cells, there is a diversity of transfer learning performance, which indicates that CNNs are cell-specific. Moreover, when CNNs are transferred between different types of input images, here white noise v.s. natural images, transfer learning shows a good performance, which implies that CNN indeed captures the full computational ability of a single retinal cell for different inputs. Taken together, these results suggest that CNN could be used to reveal structure components of neuronal circuits, and provide a powerful model for neural system identification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge