Machine-Learning-Based Diagnostics of EEG Pathology
Paper and Code
Feb 11, 2020
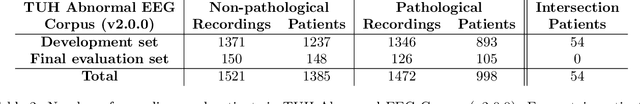
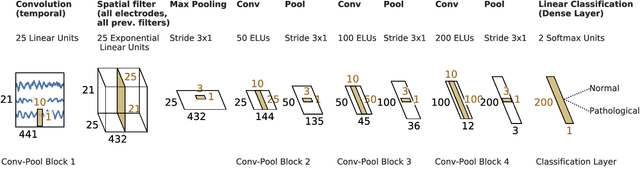
Machine learning (ML) methods have the potential to automate clinical EEG analysis. They can be categorized into feature-based (with handcrafted features), and end-to-end approaches (with learned features). Previous studies on EEG pathology decoding have typically analyzed a limited number of features, decoders, or both. For a I) more elaborate feature-based EEG analysis, and II) in-depth comparisons of both approaches, here we first develop a comprehensive feature-based framework, and then compare this framework to state-of-the-art end-to-end methods. To this aim, we apply the proposed feature-based framework and deep neural networks including an EEG-optimized temporal convolutional network (TCN) to the task of pathological versus non-pathological EEG classification. For a robust comparison, we chose the Temple University Hospital (TUH) Abnormal EEG Corpus (v2.0.0), which contains approximately 3000 EEG recordings. The results demonstrate that the proposed feature-based decoding framework can achieve accuracies on the same level as state-of-the-art deep neural networks. We find accuracies across both approaches in an astonishingly narrow range from 81--86\%. Moreover, visualizations and analyses indicated that both approaches used similar aspects of the data, e.g., delta and theta band power at temporal electrode locations. We argue that the accuracies of current binary EEG pathology decoders could saturate near 90\% due to the imperfect inter-rater agreement of the clinical labels, and that such decoders are already clinically useful, such as in areas where clinical EEG experts are rare. We make the proposed feature-based framework available open source and thus offer a new tool for EEG machine learning research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge