Zhongliang Zhou
DiffusionQC: Artifact Detection in Histopathology via Diffusion Model
Jan 18, 2026Abstract:Digital pathology plays a vital role across modern medicine, offering critical insights for disease diagnosis, prognosis, and treatment. However, histopathology images often contain artifacts introduced during slide preparation and digitization. Detecting and excluding them is essential to ensure reliable downstream analysis. Traditional supervised models typically require large annotated datasets, which is resource-intensive and not generalizable to novel artifact types. To address this, we propose DiffusionQC, which detects artifacts as outliers among clean images using a diffusion model. It requires only a set of clean images for training rather than pixel-level artifact annotations and predefined artifact types. Furthermore, we introduce a contrastive learning module to explicitly enlarge the distribution separation between artifact and clean images, yielding an enhanced version of our method. Empirical results demonstrate superior performance to state-of-the-art and offer cross-stain generalization capacity, with significantly less data and annotations.
Enhanced Diagnostic Performance via Large-Resolution Inference Optimization for Pathology Foundation Models
Jan 17, 2026Abstract:Despite their prominent performance on tasks such as ROI classification and segmentation, many pathology foundation models remain constrained by a specific input size e.g. 224 x 224, creating substantial inefficiencies when applied to whole-slide images (WSIs), which span thousands of resolutions. A naive strategy is to either enlarge inputs or downsample the WSIs. However, enlarging inputs results in prohibitive GPU memory consumption, while downsampling alters the microns-per-pixel resolution and obscures critical morphological details. To overcome these limitations, we propose an space- and time- efficient inference strategy that sparsifies attention using spatially aware neighboring blocks and filters out non-informative tokens through global attention scores. This design substantially reduces GPU memory and runtime during high-resolution WSI inference while preserving and even improving the downstream performance, enabling inference at higher resolutions under the same GPU budget. The experimental results show that our method can achieves up to an 7.67% improvement in the ROI classification and compatible results in segmentation.
Img2Loc: Revisiting Image Geolocalization using Multi-modality Foundation Models and Image-based Retrieval-Augmented Generation
Mar 28, 2024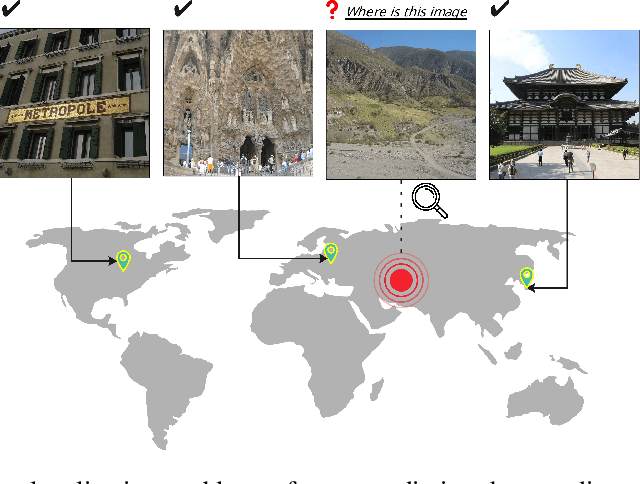

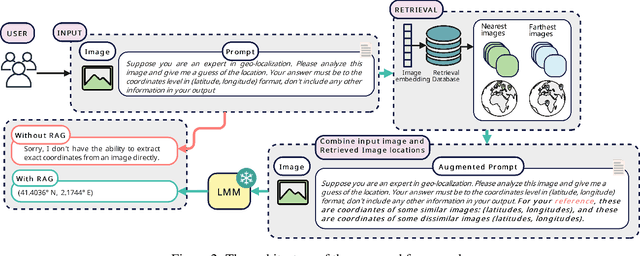
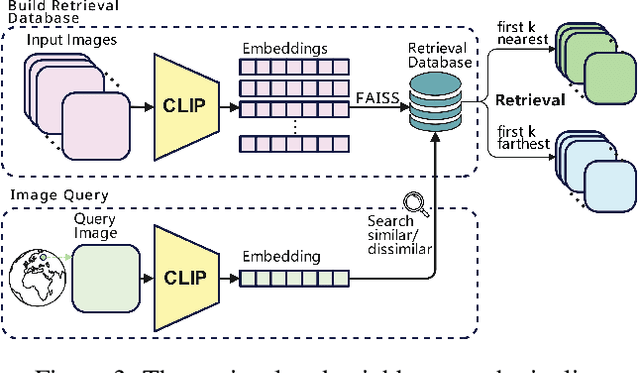
Abstract:Geolocating precise locations from images presents a challenging problem in computer vision and information retrieval.Traditional methods typically employ either classification, which dividing the Earth surface into grid cells and classifying images accordingly, or retrieval, which identifying locations by matching images with a database of image-location pairs. However, classification-based approaches are limited by the cell size and cannot yield precise predictions, while retrieval-based systems usually suffer from poor search quality and inadequate coverage of the global landscape at varied scale and aggregation levels. To overcome these drawbacks, we present Img2Loc, a novel system that redefines image geolocalization as a text generation task. This is achieved using cutting-edge large multi-modality models like GPT4V or LLaVA with retrieval augmented generation. Img2Loc first employs CLIP-based representations to generate an image-based coordinate query database. It then uniquely combines query results with images itself, forming elaborate prompts customized for LMMs. When tested on benchmark datasets such as Im2GPS3k and YFCC4k, Img2Loc not only surpasses the performance of previous state-of-the-art models but does so without any model training.
XAI meets Biology: A Comprehensive Review of Explainable AI in Bioinformatics Applications
Dec 11, 2023

Abstract:Artificial intelligence (AI), particularly machine learning and deep learning models, has significantly impacted bioinformatics research by offering powerful tools for analyzing complex biological data. However, the lack of interpretability and transparency of these models presents challenges in leveraging these models for deeper biological insights and for generating testable hypotheses. Explainable AI (XAI) has emerged as a promising solution to enhance the transparency and interpretability of AI models in bioinformatics. This review provides a comprehensive analysis of various XAI techniques and their applications across various bioinformatics domains including DNA, RNA, and protein sequence analysis, structural analysis, gene expression and genome analysis, and bioimaging analysis. We introduce the most pertinent machine learning and XAI methods, then discuss their diverse applications and address the current limitations of available XAI tools. By offering insights into XAI's potential and challenges, this review aims to facilitate its practical implementation in bioinformatics research and help researchers navigate the landscape of XAI tools.
BadSAM: Exploring Security Vulnerabilities of SAM via Backdoor Attacks
May 05, 2023Abstract:Recently, the Segment Anything Model (SAM) has gained significant attention as an image segmentation foundation model due to its strong performance on various downstream tasks. However, it has been found that SAM does not always perform satisfactorily when faced with challenging downstream tasks. This has led downstream users to demand a customized SAM model that can be adapted to these downstream tasks. In this paper, we present BadSAM, the first backdoor attack on the image segmentation foundation model. Our preliminary experiments on the CAMO dataset demonstrate the effectiveness of BadSAM.
Text2Seg: Remote Sensing Image Semantic Segmentation via Text-Guided Visual Foundation Models
Apr 20, 2023Abstract:Recent advancements in foundation models (FMs), such as GPT-4 and LLaMA, have attracted significant attention due to their exceptional performance in zero-shot learning scenarios. Similarly, in the field of visual learning, models like Grounding DINO and the Segment Anything Model (SAM) have exhibited remarkable progress in open-set detection and instance segmentation tasks. It is undeniable that these FMs will profoundly impact a wide range of real-world visual learning tasks, ushering in a new paradigm shift for developing such models. In this study, we concentrate on the remote sensing domain, where the images are notably dissimilar from those in conventional scenarios. We developed a pipeline that leverages multiple FMs to facilitate remote sensing image semantic segmentation tasks guided by text prompt, which we denote as Text2Seg. The pipeline is benchmarked on several widely-used remote sensing datasets, and we present preliminary results to demonstrate its effectiveness. Through this work, we aim to provide insights into maximizing the applicability of visual FMs in specific contexts with minimal model tuning. The code is available at https://github.com/Douglas2Code/Text2Seg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge