Zhi Wen
Towards Trustworthy Automatic Diagnosis Systems by Emulating Doctors' Reasoning with Deep Reinforcement Learning
Oct 13, 2022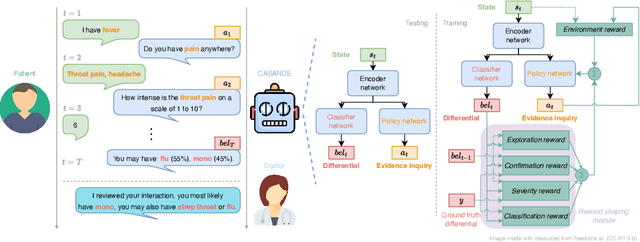
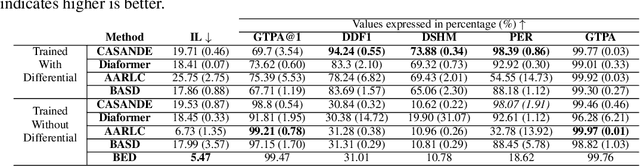
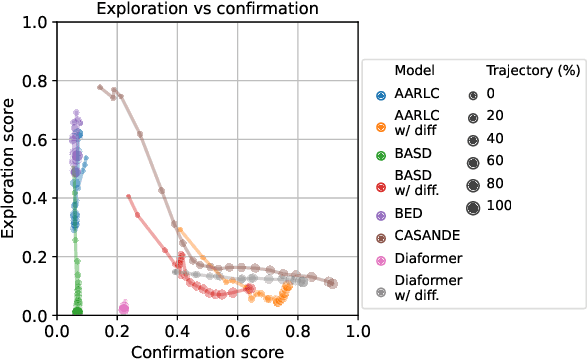
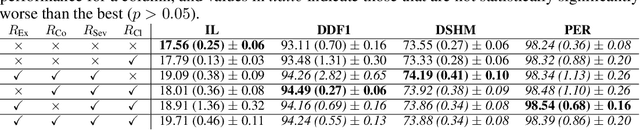
Abstract:The automation of the medical evidence acquisition and diagnosis process has recently attracted increasing attention in order to reduce the workload of doctors and democratize access to medical care. However, most works proposed in the machine learning literature focus solely on improving the prediction accuracy of a patient's pathology. We argue that this objective is insufficient to ensure doctors' acceptability of such systems. In their initial interaction with patients, doctors do not only focus on identifying the pathology a patient is suffering from; they instead generate a differential diagnosis (in the form of a short list of plausible diseases) because the medical evidence collected from patients is often insufficient to establish a final diagnosis. Moreover, doctors explicitly explore severe pathologies before potentially ruling them out from the differential, especially in acute care settings. Finally, for doctors to trust a system's recommendations, they need to understand how the gathered evidences led to the predicted diseases. In particular, interactions between a system and a patient need to emulate the reasoning of doctors. We therefore propose to model the evidence acquisition and automatic diagnosis tasks using a deep reinforcement learning framework that considers three essential aspects of a doctor's reasoning, namely generating a differential diagnosis using an exploration-confirmation approach while prioritizing severe pathologies. We propose metrics for evaluating interaction quality based on these three aspects. We show that our approach performs better than existing models while maintaining competitive pathology prediction accuracy.
DDXPlus: A new Dataset for Medical Automatic Diagnosis
May 18, 2022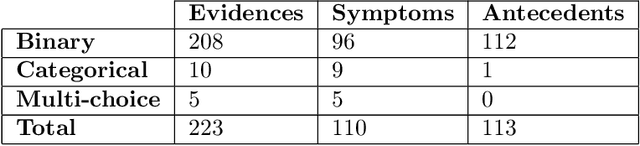
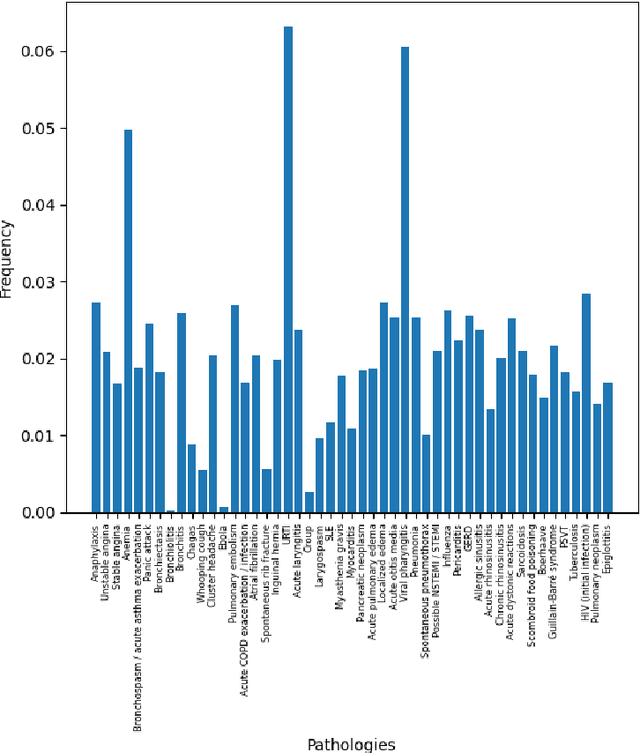
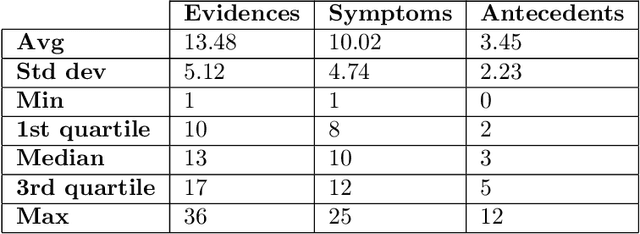
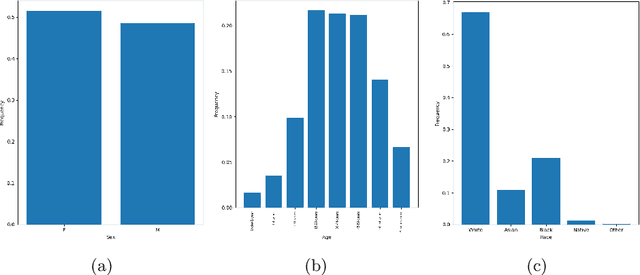
Abstract:There has been rapidly growing interests in Automatic Diagnosis (AD) and Automatic Symptom Detection (ASD) systems in the machine learning research literature, aiming to assist doctors in telemedicine services. These systems are designed to interact with patients, collect evidence relevant to their concerns, and make predictions about the underlying diseases. Doctors would review the interaction, including the evidence and the predictions, before making their final decisions. Despite the recent progress, an important piece of doctors' interactions with patients is missing in the design of AD and ASD systems, namely the differential diagnosis. Its absence is largely due to the lack of datasets that include such information for models to train on. In this work, we present a large-scale synthetic dataset that includes a differential diagnosis, along with the ground truth pathology, for each patient. In addition, this dataset includes more pathologies, as well as types of symtoms and antecedents. As a proof-of-concept, we extend several existing AD and ASD systems to incorporate differential diagnosis, and provide empirical evidence that using differentials in training signals is essential for such systems to learn to predict differentials. Dataset available at https://github.com/bruzwen/ddxplus
MeDAL: Medical Abbreviation Disambiguation Dataset for Natural Language Understanding Pretraining
Dec 27, 2020



Abstract:One of the biggest challenges that prohibit the use of many current NLP methods in clinical settings is the availability of public datasets. In this work, we present MeDAL, a large medical text dataset curated for abbreviation disambiguation, designed for natural language understanding pre-training in the medical domain. We pre-trained several models of common architectures on this dataset and empirically showed that such pre-training leads to improved performance and convergence speed when fine-tuning on downstream medical tasks.
* EMNLP 2020 Clinical NLP
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge