Zhanqiang Guo
Neuro-3D: Towards 3D Visual Decoding from EEG Signals
Nov 21, 2024Abstract:Human's perception of the visual world is shaped by the stereo processing of 3D information. Understanding how the brain perceives and processes 3D visual stimuli in the real world has been a longstanding endeavor in neuroscience. Towards this goal, we introduce a new neuroscience task: decoding 3D visual perception from EEG signals, a neuroimaging technique that enables real-time monitoring of neural dynamics enriched with complex visual cues. To provide the essential benchmark, we first present EEG-3D, a pioneering dataset featuring multimodal analysis data and extensive EEG recordings from 12 subjects viewing 72 categories of 3D objects rendered in both videos and images. Furthermore, we propose Neuro-3D, a 3D visual decoding framework based on EEG signals. This framework adaptively integrates EEG features derived from static and dynamic stimuli to learn complementary and robust neural representations, which are subsequently utilized to recover both the shape and color of 3D objects through the proposed diffusion-based colored point cloud decoder. To the best of our knowledge, we are the first to explore EEG-based 3D visual decoding. Experiments indicate that Neuro-3D not only reconstructs colored 3D objects with high fidelity, but also learns effective neural representations that enable insightful brain region analysis. The dataset and associated code will be made publicly available.
3D Vascular Segmentation Supervised by 2D Annotation of Maximum Intensity Projection
Feb 19, 2024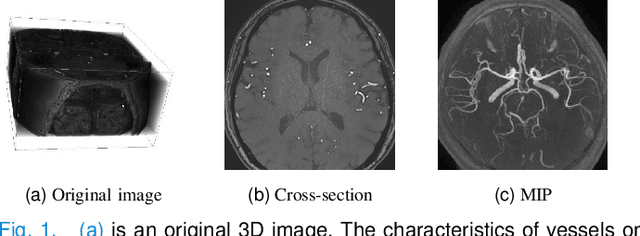



Abstract:Vascular structure segmentation plays a crucial role in medical analysis and clinical applications. The practical adoption of fully supervised segmentation models is impeded by the intricacy and time-consuming nature of annotating vessels in the 3D space. This has spurred the exploration of weakly-supervised approaches that reduce reliance on expensive segmentation annotations. Despite this, existing weakly supervised methods employed in organ segmentation, which encompass points, bounding boxes, or graffiti, have exhibited suboptimal performance when handling sparse vascular structure. To alleviate this issue, we employ maximum intensity projection (MIP) to decrease the dimensionality of 3D volume to 2D image for efficient annotation, and the 2D labels are utilized to provide guidance and oversight for training 3D vessel segmentation model. Initially, we generate pseudo-labels for 3D blood vessels using the annotations of 2D projections. Subsequently, taking into account the acquisition method of the 2D labels, we introduce a weakly-supervised network that fuses 2D-3D deep features via MIP to further improve segmentation performance. Furthermore, we integrate confidence learning and uncertainty estimation to refine the generated pseudo-labels, followed by fine-tuning the segmentation network. Our method is validated on five datasets (including cerebral vessel, aorta and coronary artery), demonstrating highly competitive performance in segmenting vessels and the potential to significantly reduce the time and effort required for vessel annotation. Our code is available at: https://github.com/gzq17/Weakly-Supervised-by-MIP.
Cerebrovascular Segmentation via Vessel Oriented Filtering Network
Oct 17, 2022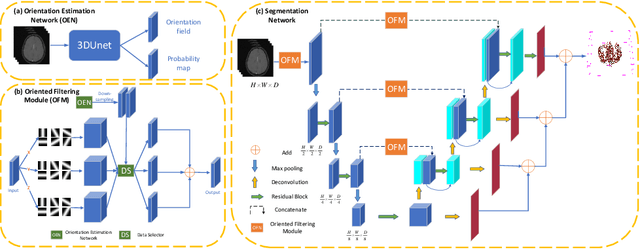
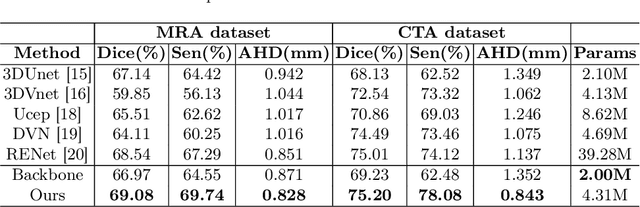
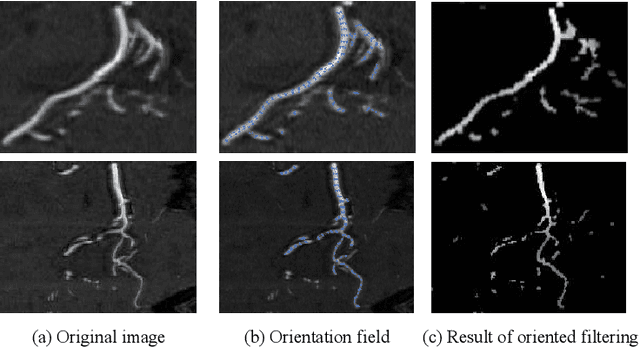

Abstract:Accurate cerebrovascular segmentation from Magnetic Resonance Angiography (MRA) and Computed Tomography Angiography (CTA) is of great significance in diagnosis and treatment of cerebrovascular pathology. Due to the complexity and topology variability of blood vessels, complete and accurate segmentation of vascular network is still a challenge. In this paper, we proposed a Vessel Oriented Filtering Network (VOF-Net) which embeds domain knowledge into the convolutional neural network. We design oriented filters for blood vessels according to vessel orientation field, which is obtained by orientation estimation network. Features extracted by oriented filtering are injected into segmentation network, so as to make use of the prior information that the blood vessels are slender and curved tubular structure. Experimental results on datasets of CTA and MRA show that the proposed method is effective for vessel segmentation, and embedding the specific vascular filter improves the segmentation performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge