Zhangxin Chen
Low Rank Optimization for Efficient Deep Learning: Making A Balance between Compact Architecture and Fast Training
Mar 22, 2023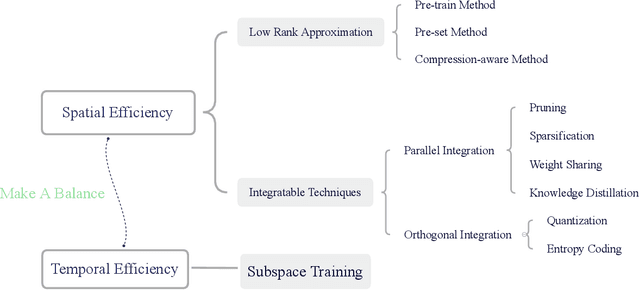
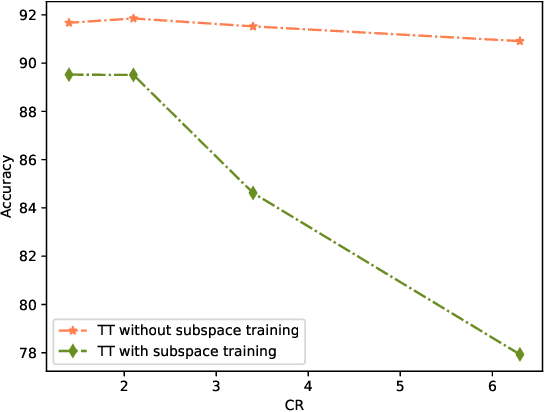
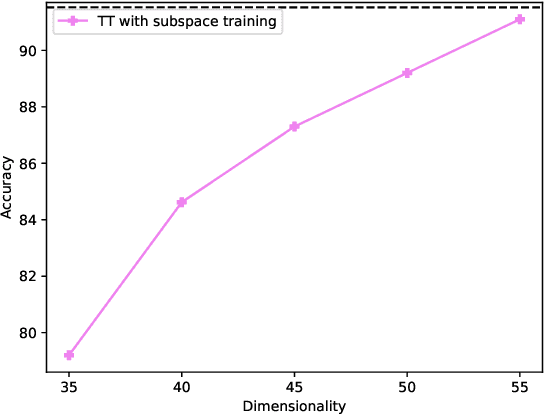
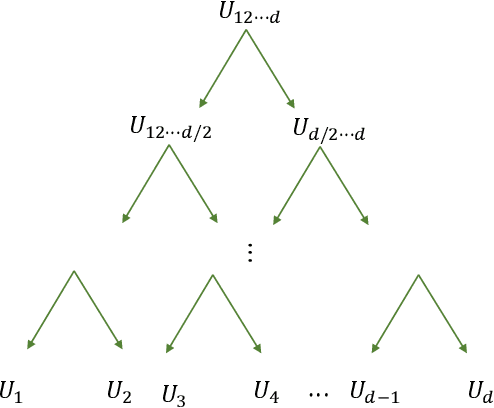
Abstract:Deep neural networks have achieved great success in many data processing applications. However, the high computational complexity and storage cost makes deep learning hard to be used on resource-constrained devices, and it is not environmental-friendly with much power cost. In this paper, we focus on low-rank optimization for efficient deep learning techniques. In the space domain, deep neural networks are compressed by low rank approximation of the network parameters, which directly reduces the storage requirement with a smaller number of network parameters. In the time domain, the network parameters can be trained in a few subspaces, which enables efficient training for fast convergence. The model compression in the spatial domain is summarized into three categories as pre-train, pre-set, and compression-aware methods, respectively. With a series of integrable techniques discussed, such as sparse pruning, quantization, and entropy coding, we can ensemble them in an integration framework with lower computational complexity and storage. Besides of summary of recent technical advances, we have two findings for motivating future works: one is that the effective rank outperforms other sparse measures for network compression. The other is a spatial and temporal balance for tensorized neural networks.
Tucker-O-Minus Decomposition for Multi-view Tensor Subspace Clustering
Oct 23, 2022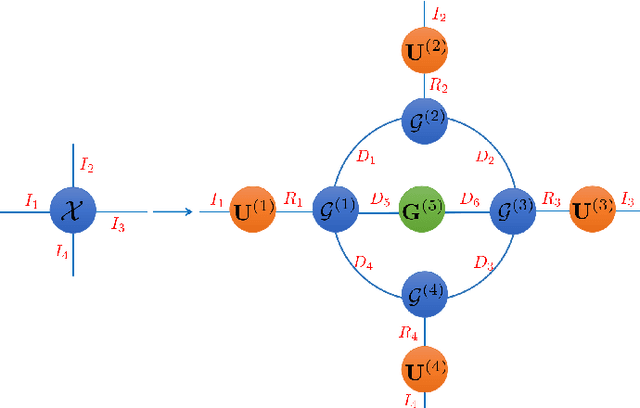
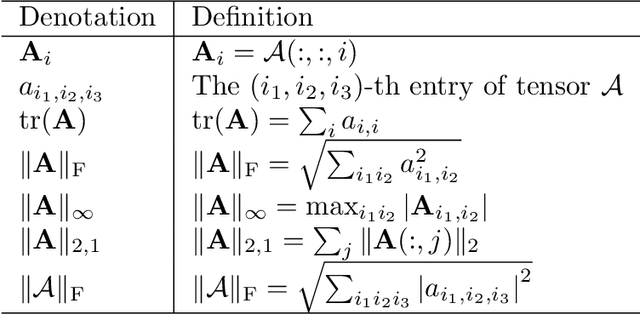
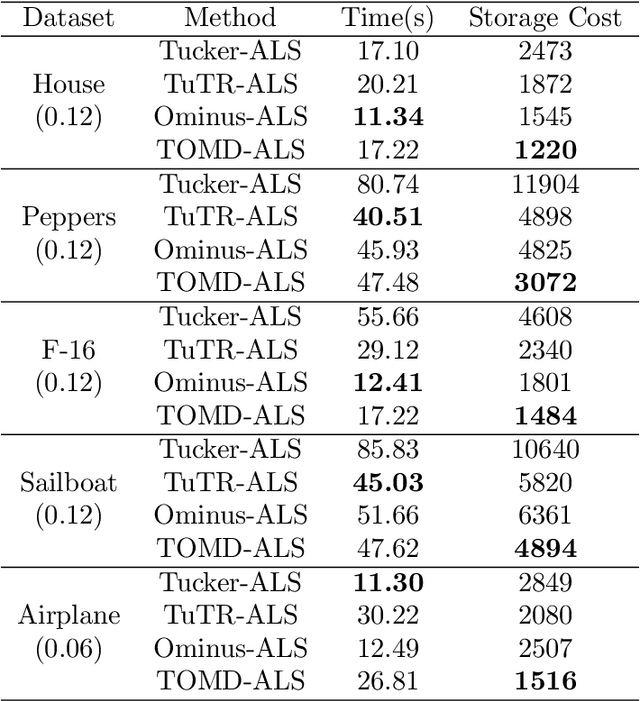
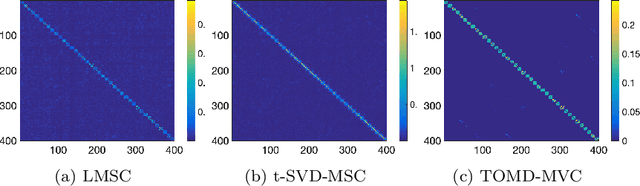
Abstract:With powerful ability to exploit latent structure of self-representation information, different tensor decompositions have been employed into low rank multi-view clustering (LRMVC) models for achieving significant performance. However, current approaches suffer from a series of problems related to those tensor decomposition, such as the unbalanced matricization scheme, rotation sensitivity, deficient correlations capture and so forth. All these will lead to LRMVC having insufficient access to global information, which is contrary to the target of multi-view clustering. To alleviate these problems, we propose a new tensor decomposition called Tucker-O-Minus Decomposition (TOMD) for multi-view clustering. Specifically, based on the Tucker format, we additionally employ the O-minus structure, which consists of a circle with an efficient bridge linking two weekly correlated factors. In this way, the core tensor in Tucker format is replaced by the O-minus architecture with a more balanced structure, and the enhanced capacity of capturing the global low rank information will be achieved. The proposed TOMD also provides more compact and powerful representation abilities for the self-representation tensor, simultaneously. The alternating direction method of multipliers is used to solve the proposed model TOMD-MVC. Numerical experiments on six benchmark data sets demonstrate the superiority of our proposed method in terms of F-score, precision, recall, normalized mutual information, adjusted rand index, and accuracy.
ProLanGO: Protein Function Prediction Using Neural~Machine Translation Based on a Recurrent Neural Network
Oct 19, 2017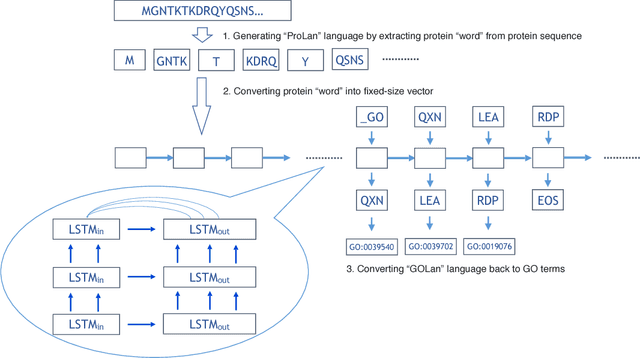
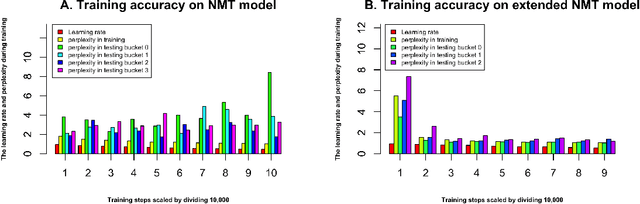
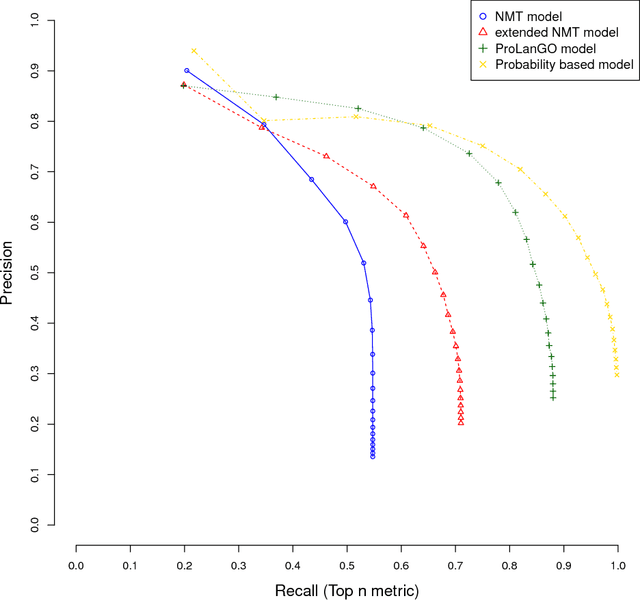
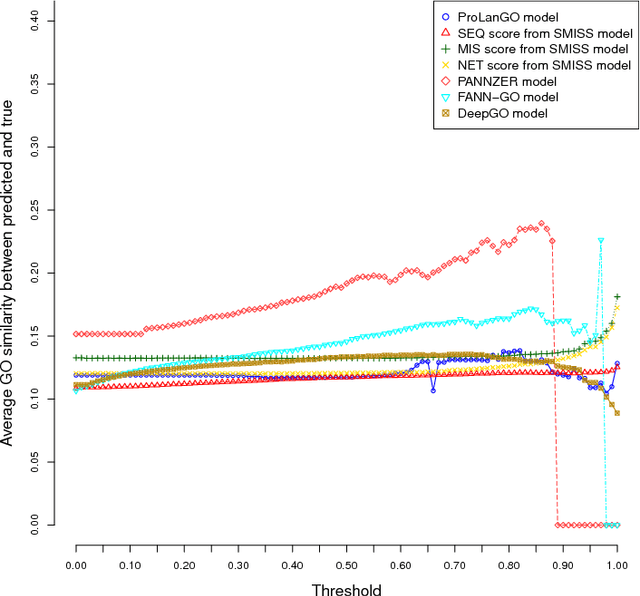
Abstract:With the development of next generation sequencing techniques, it is fast and cheap to determine protein sequences but relatively slow and expensive to extract useful information from protein sequences because of limitations of traditional biological experimental techniques. Protein function prediction has been a long standing challenge to fill the gap between the huge amount of protein sequences and the known function. In this paper, we propose a novel method to convert the protein function problem into a language translation problem by the new proposed protein sequence language "ProLan" to the protein function language "GOLan", and build a neural machine translation model based on recurrent neural networks to translate "ProLan" language to "GOLan" language. We blindly tested our method by attending the latest third Critical Assessment of Function Annotation (CAFA 3) in 2016, and also evaluate the performance of our methods on selected proteins whose function was released after CAFA competition. The good performance on the training and testing datasets demonstrates that our new proposed method is a promising direction for protein function prediction. In summary, we first time propose a method which converts the protein function prediction problem to a language translation problem and applies a neural machine translation model for protein function prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge