Zachary MC Baum
Rapid Lung Ultrasound COVID-19 Severity Scoring with Resource-Efficient Deep Feature Extraction
Jul 22, 2022
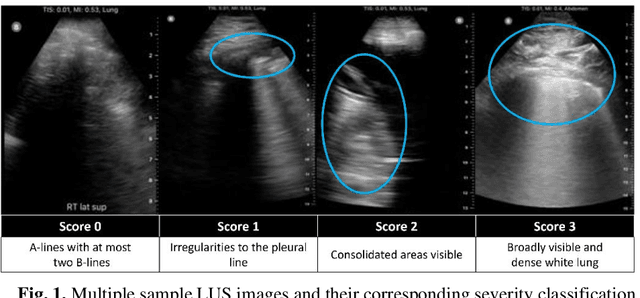
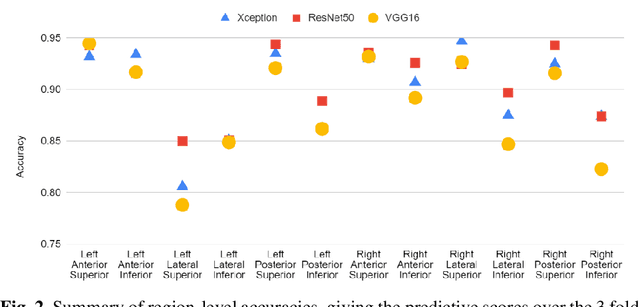
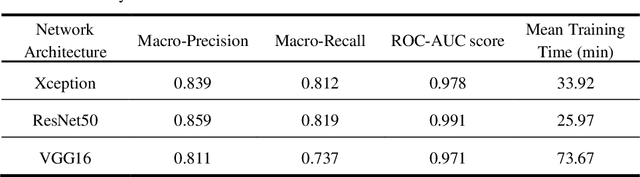
Abstract:Artificial intelligence-based analysis of lung ultrasound imaging has been demonstrated as an effective technique for rapid diagnostic decision support throughout the COVID-19 pandemic. However, such techniques can require days- or weeks-long training processes and hyper-parameter tuning to develop intelligent deep learning image analysis models. This work focuses on leveraging 'off-the-shelf' pre-trained models as deep feature extractors for scoring disease severity with minimal training time. We propose using pre-trained initializations of existing methods ahead of simple and compact neural networks to reduce reliance on computational capacity. This reduction of computational capacity is of critical importance in time-limited or resource-constrained circumstances, such as the early stages of a pandemic. On a dataset of 49 patients, comprising over 20,000 images, we demonstrate that the use of existing methods as feature extractors results in the effective classification of COVID-19-related pneumonia severity while requiring only minutes of training time. Our methods can achieve an accuracy of over 0.93 on a 4-level severity score scale and provides comparable per-patient region and global scores compared to expert annotated ground truths. These results demonstrate the capability for rapid deployment and use of such minimally-adapted methods for progress monitoring, patient stratification and management in clinical practice for COVID-19 patients, and potentially in other respiratory diseases.
Meta-Registration: Learning Test-Time Optimization for Single-Pair Image Registration
Jul 22, 2022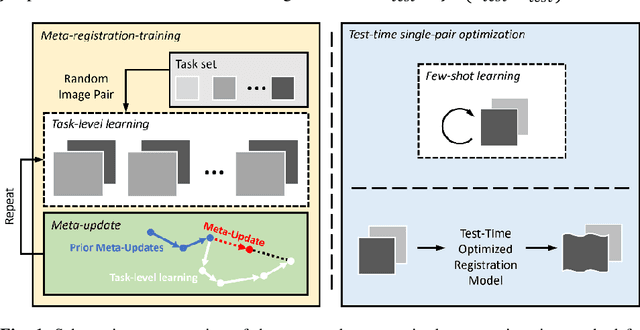



Abstract:Neural networks have been proposed for medical image registration by learning, with a substantial amount of training data, the optimal transformations between image pairs. These trained networks can further be optimized on a single pair of test images - known as test-time optimization. This work formulates image registration as a meta-learning algorithm. Such networks can be trained by aligning the training image pairs while simultaneously improving test-time optimization efficacy; tasks which were previously considered two independent training and optimization processes. The proposed meta-registration is hypothesized to maximize the efficiency and effectiveness of the test-time optimization in the "outer" meta-optimization of the networks. For image guidance applications that often are time-critical yet limited in training data, the potentially gained speed and accuracy are compared with classical registration algorithms, registration networks without meta-learning, and single-pair optimization without test-time optimization data. Experiments are presented in this paper using clinical transrectal ultrasound image data from 108 prostate cancer patients. These experiments demonstrate the effectiveness of a meta-registration protocol, which yields significantly improved performance relative to existing learning-based methods. Furthermore, the meta-registration achieves comparable results to classical iterative methods in a fraction of the time, owing to its rapid test-time optimization process.
Learning Generalized Non-Rigid Multimodal Biomedical Image Registration from Generic Point Set Data
Jul 22, 2022



Abstract:Free Point Transformer (FPT) has been proposed as a data-driven, non-rigid point set registration approach using deep neural networks. As FPT does not assume constraints based on point vicinity or correspondence, it may be trained simply and in a flexible manner by minimizing an unsupervised loss based on the Chamfer Distance. This makes FPT amenable to real-world medical imaging applications where ground-truth deformations may be infeasible to obtain, or in scenarios where only a varying degree of completeness in the point sets to be aligned is available. To test the limit of the correspondence finding ability of FPT and its dependency on training data sets, this work explores the generalizability of the FPT from well-curated non-medical data sets to medical imaging data sets. First, we train FPT on the ModelNet40 dataset to demonstrate its effectiveness and the superior registration performance of FPT over iterative and learning-based point set registration methods. Second, we demonstrate superior performance in rigid and non-rigid registration and robustness to missing data. Last, we highlight the interesting generalizability of the ModelNet-trained FPT by registering reconstructed freehand ultrasound scans of the spine and generic spine models without additional training, whereby the average difference to the ground truth curvatures is 1.3 degrees, across 13 patients.
Lung Ultrasound Segmentation and Adaptation between COVID-19 and Community-Acquired Pneumonia
Aug 06, 2021
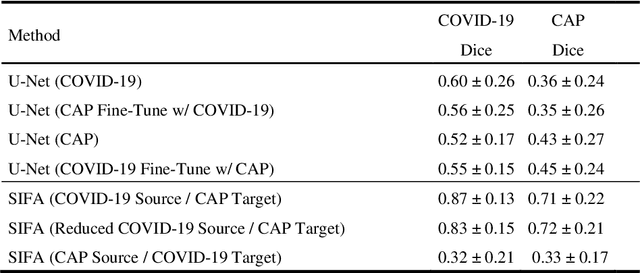
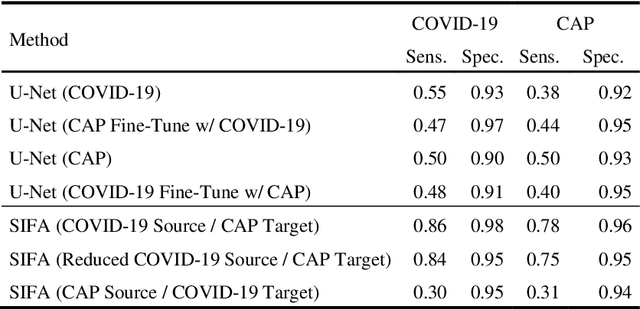
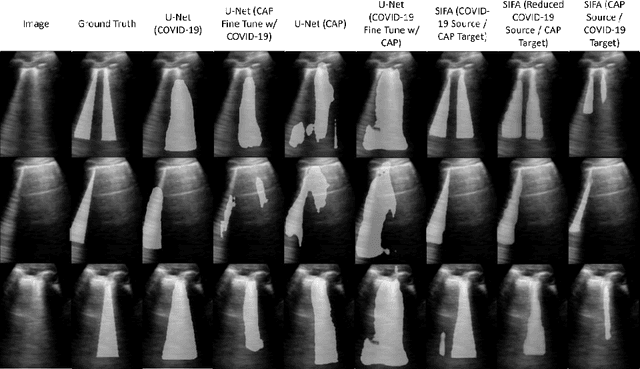
Abstract:Lung ultrasound imaging has been shown effective in detecting typical patterns for interstitial pneumonia, as a point-of-care tool for both patients with COVID-19 and other community-acquired pneumonia (CAP). In this work, we focus on the hyperechoic B-line segmentation task. Using deep neural networks, we automatically outline the regions that are indicative of pathology-sensitive artifacts and their associated sonographic patterns. With a real-world data-scarce scenario, we investigate approaches to utilize both COVID-19 and CAP lung ultrasound data to train the networks; comparing fine-tuning and unsupervised domain adaptation. Segmenting either type of lung condition at inference may support a range of clinical applications during evolving epidemic stages, but also demonstrates value in resource-constrained clinical scenarios. Adapting real clinical data acquired from COVID-19 patients to those from CAP patients significantly improved Dice scores from 0.60 to 0.87 (p < 0.001) and from 0.43 to 0.71 (p < 0.001), on independent COVID-19 and CAP test cases, respectively. It is of practical value that the improvement was demonstrated with only a small amount of data in both training and adaptation data sets, a common constraint for deploying machine learning models in clinical practice. Interestingly, we also report that the inverse adaptation, from labelled CAP data to unlabeled COVID-19 data, did not demonstrate an improvement when tested on either condition. Furthermore, we offer a possible explanation that correlates the segmentation performance to label consistency and data domain diversity in this point-of-care lung ultrasound application.
Development and evaluation of intraoperative ultrasound segmentation with negative image frames and multiple observer labels
Jul 28, 2021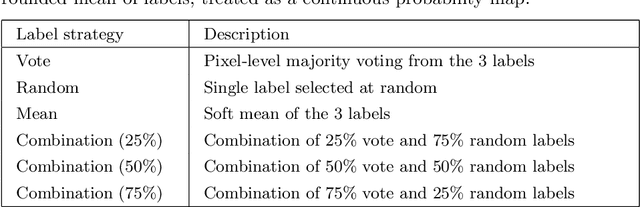
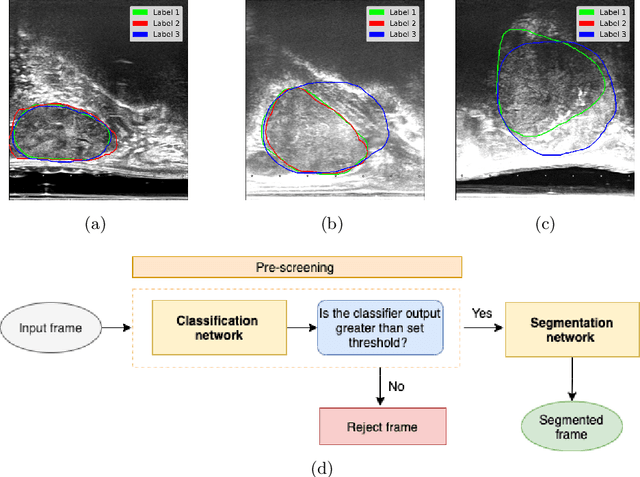
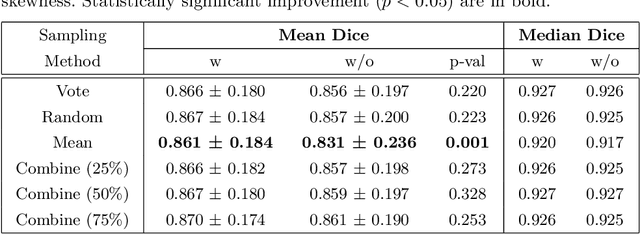
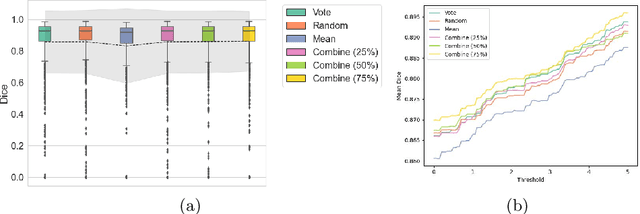
Abstract:When developing deep neural networks for segmenting intraoperative ultrasound images, several practical issues are encountered frequently, such as the presence of ultrasound frames that do not contain regions of interest and the high variance in ground-truth labels. In this study, we evaluate the utility of a pre-screening classification network prior to the segmentation network. Experimental results demonstrate that such a classifier, minimising frame classification errors, was able to directly impact the number of false positive and false negative frames. Importantly, the segmentation accuracy on the classifier-selected frames, that would be segmented, remains comparable to or better than those from standalone segmentation networks. Interestingly, the efficacy of the pre-screening classifier was affected by the sampling methods for training labels from multiple observers, a seemingly independent problem. We show experimentally that a previously proposed approach, combining random sampling and consensus labels, may need to be adapted to perform well in our application. Furthermore, this work aims to share practical experience in developing a machine learning application that assists highly variable interventional imaging for prostate cancer patients, to present robust and reproducible open-source implementations, and to report a set of comprehensive results and analysis comparing these practical, yet important, options in a real-world clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge