Yvonne A. Evrard
Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, Inc. Frederick, MD, USA
Learning Curves for Drug Response Prediction in Cancer Cell Lines
Nov 25, 2020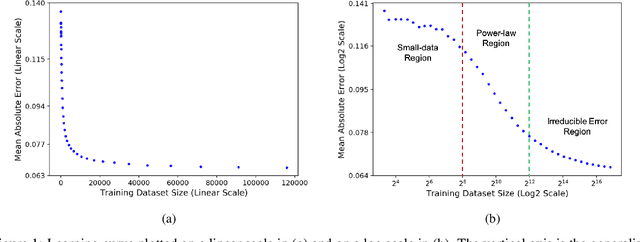
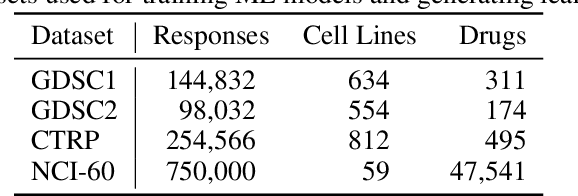
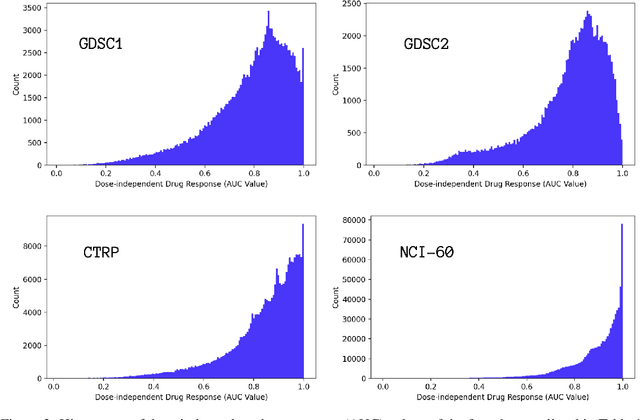
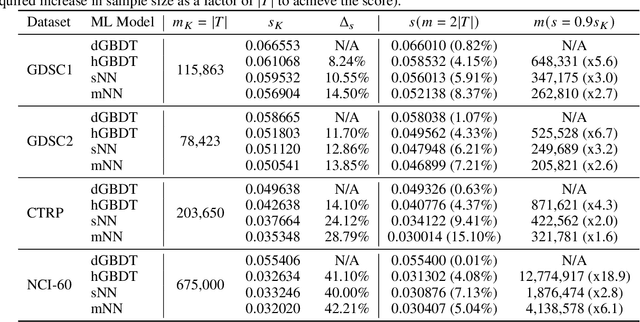
Abstract:Motivated by the size of cell line drug sensitivity data, researchers have been developing machine learning (ML) models for predicting drug response to advance cancer treatment. As drug sensitivity studies continue generating data, a common question is whether the proposed predictors can further improve the generalization performance with more training data. We utilize empirical learning curves for evaluating and comparing the data scaling properties of two neural networks (NNs) and two gradient boosting decision tree (GBDT) models trained on four drug screening datasets. The learning curves are accurately fitted to a power law model, providing a framework for assessing the data scaling behavior of these predictors. The curves demonstrate that no single model dominates in terms of prediction performance across all datasets and training sizes, suggesting that the shape of these curves depends on the unique model-dataset pair. The multi-input NN (mNN), in which gene expressions and molecular drug descriptors are input into separate subnetworks, outperforms a single-input NN (sNN), where the cell and drug features are concatenated for the input layer. In contrast, a GBDT with hyperparameter tuning exhibits superior performance as compared with both NNs at the lower range of training sizes for two of the datasets, whereas the mNN performs better at the higher range of training sizes. Moreover, the trajectory of the curves suggests that increasing the sample size is expected to further improve prediction scores of both NNs. These observations demonstrate the benefit of using learning curves to evaluate predictors, providing a broader perspective on the overall data scaling characteristics. The fitted power law curves provide a forward-looking performance metric and can serve as a co-design tool to guide experimental biologists and computational scientists in the design of future experiments.
Ensemble Transfer Learning for the Prediction of Anti-Cancer Drug Response
May 13, 2020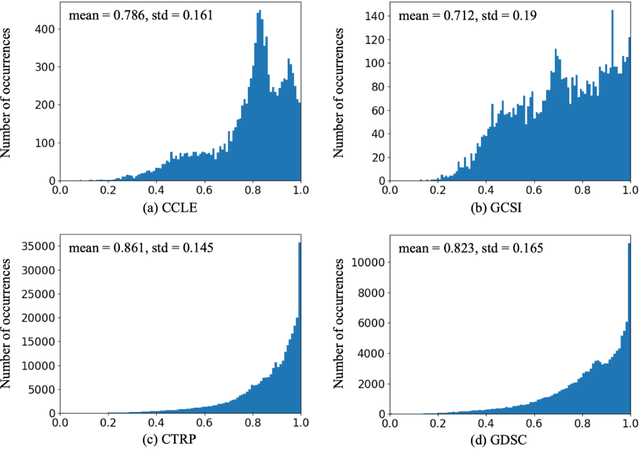
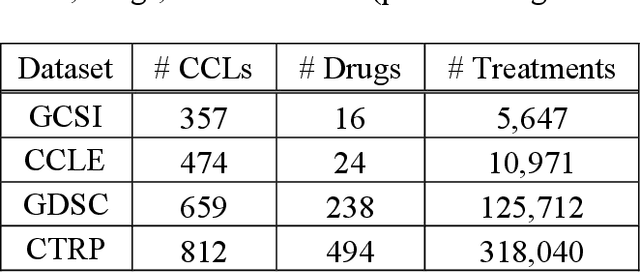
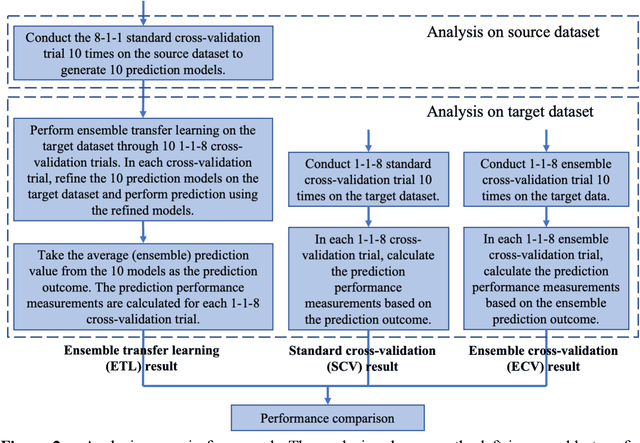
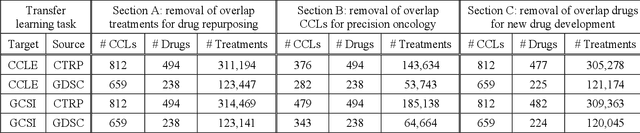
Abstract:Transfer learning has been shown to be effective in many applications in which training data for the target problem are limited but data for a related (source) problem are abundant. In this paper, we apply transfer learning to the prediction of anti-cancer drug response. Previous transfer learning studies for drug response prediction focused on building models that predict the response of tumor cells to a specific drug treatment. We target the more challenging task of building general prediction models that can make predictions for both new tumor cells and new drugs. We apply the classic transfer learning framework that trains a prediction model on the source dataset and refines it on the target dataset, and extends the framework through ensemble. The ensemble transfer learning pipeline is implemented using LightGBM and two deep neural network (DNN) models with different architectures. Uniquely, we investigate its power for three application settings including drug repurposing, precision oncology, and new drug development, through different data partition schemes in cross-validation. We test the proposed ensemble transfer learning on benchmark in vitro drug screening datasets, taking one dataset as the source domain and another dataset as the target domain. The analysis results demonstrate the benefit of applying ensemble transfer learning for predicting anti-cancer drug response in all three applications with both LightGBM and DNN models. Compared between the different prediction models, a DNN model with two subnetworks for the inputs of tumor features and drug features separately outperforms LightGBM and the other DNN model that concatenates tumor features and drug features for input in the drug repurposing and precision oncology applications. In the more challenging application of new drug development, LightGBM performs better than the other two DNN models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge