Yufan Luo
G-RCN: Optimizing the Gap between Classification and Localization Tasks for Object Detection
Nov 14, 2020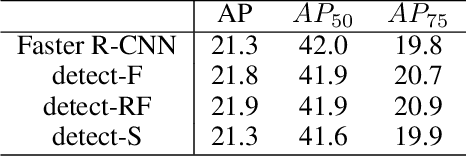
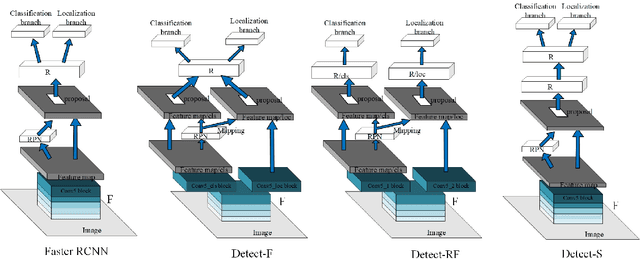
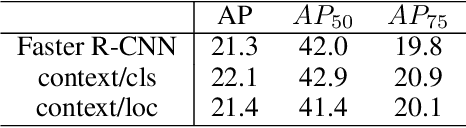
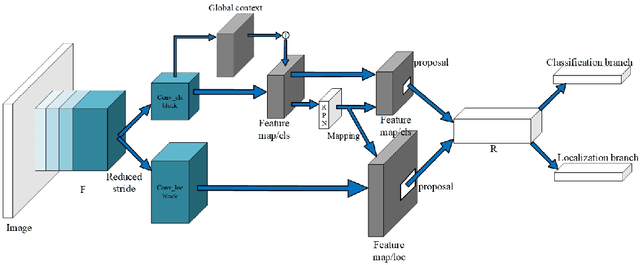
Abstract:Multi-task learning is widely used in computer vision. Currently, object detection models utilize shared feature map to complete classification and localization tasks simultaneously. By comparing the performance between the original Faster R-CNN and that with partially separated feature maps, we show that: (1) Sharing high-level features for the classification and localization tasks is sub-optimal; (2) Large stride is beneficial for classification but harmful for localization; (3) Global context information could improve the performance of classification. Based on these findings, we proposed a paradigm called Gap-optimized region based convolutional network (G-RCN), which aims to separating these two tasks and optimizing the gap between them. The paradigm was firstly applied to correct the current ResNet protocol by simply reducing the stride and moving the Conv5 block from the head to the feature extraction network, which brings 3.6 improvement of AP70 on the PASCAL VOC dataset and 1.5 improvement of AP on the COCO dataset for ResNet50. Next, the new method is applied on the Faster R-CNN with backbone of VGG16,ResNet50 and ResNet101, which brings above 2.0 improvement of AP70 on the PASCAL VOC dataset and above 1.9 improvement of AP on the COCO dataset. Noticeably, the implementation of G-RCN only involves a few structural modifications, with no extra module added.
DeepACC:Automate Chromosome Classification based on Metaphase Images using Deep Learning Framework Fused with Prior Knowledge
Jun 28, 2020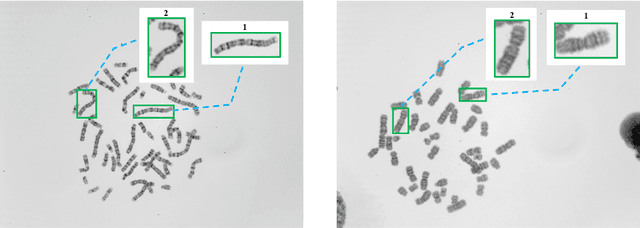
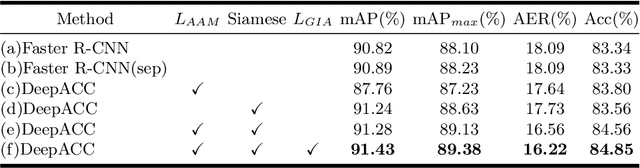
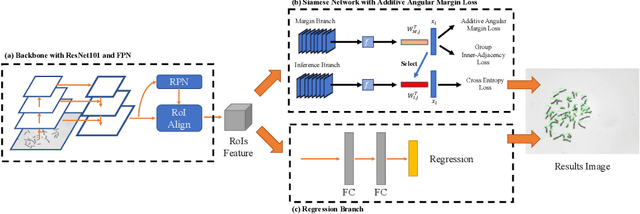

Abstract:Chromosome classification is an important but difficult and tedious task in karyotyping. Previous methods only classify manually segmented single chromosome, which is far from clinical practice. In this work, we propose a detection based method, DeepACC, to locate and fine classify chromosomes simultaneously based on the whole metaphase image. We firstly introduce the Additive Angular Margin Loss to enhance the discriminative power of model. To alleviate batch effects, we transform decision boundary of each class case-by-case through a siamese network which make full use of prior knowledges that chromosomes usually appear in pairs. Furthermore, we take the clinically seven group criterion as a prior knowledge and design an additional Group Inner-Adjacency Loss to further reduce inter-class similarities. 3390 metaphase images from clinical laboratory are collected and labelled to evaluate the performance. Results show that the new design brings encouraging performance gains comparing to the state-of-the-art baselines.
PBRnet: Pyramidal Bounding Box Refinement to Improve Object Localization Accuracy
Mar 10, 2020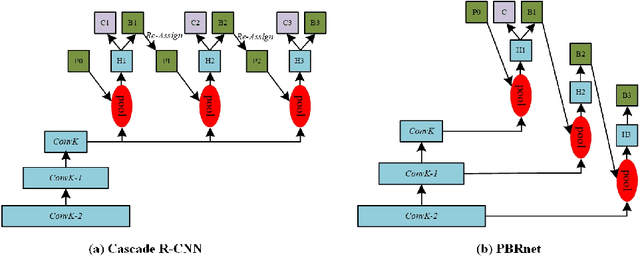
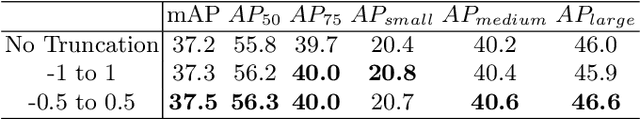
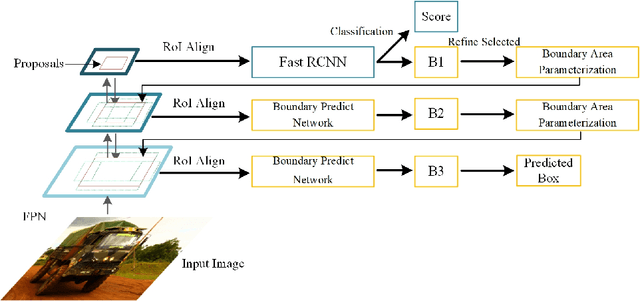
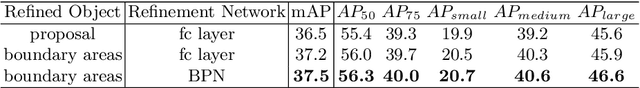
Abstract:Many recently developed object detectors focused on coarse-to-fine framework which contains several stages that classify and regress proposals from coarse-grain to fine-grain, and obtains more accurate detection gradually. Multi-resolution models such as Feature Pyramid Network(FPN) integrate information of different levels of resolution and effectively improve the performance. Previous researches also have revealed that localization can be further improved by: 1) using fine-grained information which is more translational variant; 2) refining local areas which is more focused on local boundary information. Based on these principles, we designed a novel boundary refinement architecture to improve localization accuracy by combining coarse-to-fine framework with feature pyramid structure, named as Pyramidal Bounding Box Refinement network(PBRnet), which parameterizes gradually focused boundary areas of objects and leverages lower-level feature maps to extract finer local information when refining the predicted bounding boxes. Extensive experiments are performed on the MS-COCO dataset. The PBRnet brings a significant performance gains by roughly 3 point of $mAP$ when added to FPN or Libra R-CNN. Moreover, by treating Cascade R-CNN as a coarse-to-fine detector and replacing its localization branch by the regressor of PBRnet, it leads an extra performance improvement by 1.5 $mAP$, yielding a total performance boosting by as high as 5 point of $mAP$.
DeepACE: Automated Chromosome Enumeration in Metaphase Cell Images Using Deep Convolutional Neural Networks
Oct 12, 2019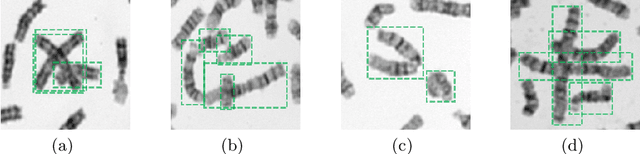
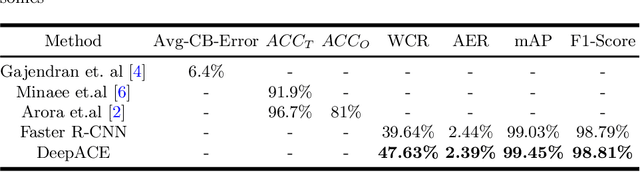
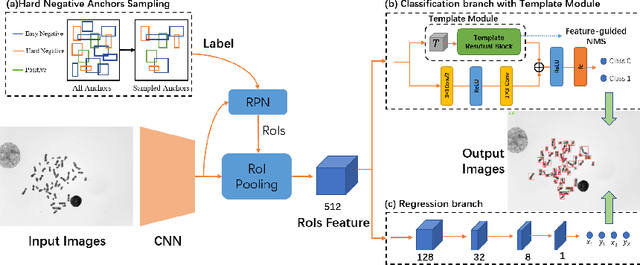
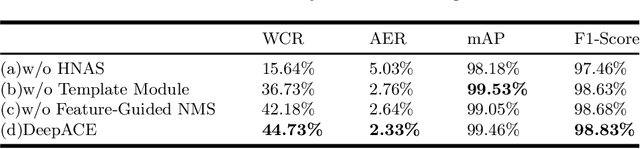
Abstract:Chromosome enumeration is an important but tedious procedure in karyotyping analysis. In this paper, to automate the enumeration process, we developed a chromosome enumeration framework, DeepACE, based on the region based object detection scheme. Firstly, the ability of region proposal network is enhanced by a newly proposed Hard Negative Anchors Sampling to extract unapparent but important information about highly confusing partial chromosomes. Next, to alleviate serious occlusion problems, we novelly introduced a weakly-supervised mechanism by adding a Template Module into classification branch to heuristically separate overlapped chromosomes. The template features are further incorporated into the NMS procedure to further improve the detection of overlapping chromosomes. In the newly collected clinical dataset, the proposed method outperform all the previous method, yielding an mAP with respect to chromosomes as 99.45, and the error rate is about 2.4%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge