Yubo Tao
MSFormer: A Skeleton-multiview Fusion Method For Tooth Instance Segmentation
Oct 23, 2023Abstract:Recently, deep learning-based tooth segmentation methods have been limited by the expensive and time-consuming processes of data collection and labeling. Achieving high-precision segmentation with limited datasets is critical. A viable solution to this entails fine-tuning pre-trained multiview-based models, thereby enhancing performance with limited data. However, relying solely on two-dimensional (2D) images for three-dimensional (3D) tooth segmentation can produce suboptimal outcomes because of occlusion and deformation, i.e., incomplete and distorted shape perception. To improve this fine-tuning-based solution, this paper advocates 2D-3D joint perception. The fundamental challenge in employing 2D-3D joint perception with limited data is that the 3D-related inputs and modules must follow a lightweight policy instead of using huge 3D data and parameter-rich modules that require extensive training data. Following this lightweight policy, this paper selects skeletons as the 3D inputs and introduces MSFormer, a novel method for tooth segmentation. MSFormer incorporates two lightweight modules into existing multiview-based models: a 3D-skeleton perception module to extract 3D perception from skeletons and a skeleton-image contrastive learning module to obtain the 2D-3D joint perception by fusing both multiview and skeleton perceptions. The experimental results reveal that MSFormer paired with large pre-trained multiview models achieves state-of-the-art performance, requiring only 100 training meshes. Furthermore, the segmentation accuracy is improved by 2.4%-5.5% with the increasing volume of training data.
voxel2vec: A Natural Language Processing Approach to Learning Distributed Representations for Scientific Data
Jul 11, 2022



Abstract:Relationships in scientific data, such as the numerical and spatial distribution relations of features in univariate data, the scalar-value combinations' relations in multivariate data, and the association of volumes in time-varying and ensemble data, are intricate and complex. This paper presents voxel2vec, a novel unsupervised representation learning model, which is used to learn distributed representations of scalar values/scalar-value combinations in a low-dimensional vector space. Its basic assumption is that if two scalar values/scalar-value combinations have similar contexts, they usually have high similarity in terms of features. By representing scalar values/scalar-value combinations as symbols, voxel2vec learns the similarity between them in the context of spatial distribution and then allows us to explore the overall association between volumes by transfer prediction. We demonstrate the usefulness and effectiveness of voxel2vec by comparing it with the isosurface similarity map of univariate data and applying the learned distributed representations to feature classification for multivariate data and to association analysis for time-varying and ensemble data.
SPAN: Subgraph Prediction Attention Network for Dynamic Graphs
Aug 17, 2021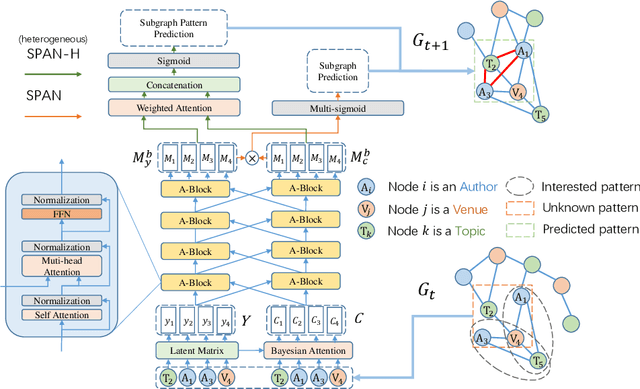



Abstract:This paper proposes a novel model for predicting subgraphs in dynamic graphs, an extension of traditional link prediction. This proposed end-to-end model learns a mapping from the subgraph structures in the current snapshot to the subgraph structures in the next snapshot directly, i.e., edge existence among multiple nodes in the subgraph. A new mechanism named cross-attention with a twin-tower module is designed to integrate node attribute information and topology information collaboratively for learning subgraph evolution. We compare our model with several state-of-the-art methods for subgraph prediction and subgraph pattern prediction in multiple real-world homogeneous and heterogeneous dynamic graphs, respectively. Experimental results demonstrate that our model outperforms other models in these two tasks, with a gain increase from 5.02% to 10.88%.
A Co-analysis Framework for Exploring Multivariate Scientific Data
Aug 19, 2019



Abstract:In complex multivariate data sets, different features usually include diverse associations with different variables, and different variables are associated within different regions. Therefore, exploring the associations between variables and voxels locally becomes necessary to better understand the underlying phenomena. In this paper, we propose a co-analysis framework based on biclusters, which are two subsets of variables and voxels with close scalar-value relationships, to guide the process of visually exploring multivariate data. We first automatically extract all meaningful biclusters, each of which only contains voxels with a similar scalar-value pattern over a subset of variables. These biclusters are organized according to their variable sets, and biclusters in each variable set are further grouped by a similarity metric to reduce redundancy and support diversity during visual exploration. Biclusters are visually represented in coordinated views to facilitate interactive exploration of multivariate data based on the similarity between biclusters and the correlation of scalar values with different variables. Experiments on several representative multivariate scientific data sets demonstrate the effectiveness of our framework in exploring local relationships among variables, biclusters and scalar values in the data.
* 31 pages, 7 figures
Multivariate Spatial Data Visualization: A Survey
Aug 18, 2019



Abstract:Multivariate spatial data plays an important role in computational science and engineering simulations. The potential features and hidden relationships in multivariate data can assist scientists to gain an in-depth understanding of a scientific process, verify a hypothesis and further discover a new physical or chemical law. In this paper, we present a comprehensive survey of the state-of-the-art techniques for multivariate spatial data visualization. We first introduce the basic concept and characteristics of multivariate spatial data, and describe three main tasks in multivariate data visualization: feature classification, fusion visualization, and correlation analysis. Finally, we prospect potential research topics for multivariate data visualization according to the current research.
* 16 pages, 5 figures. Corresponding author: Yubo Tao
Dynamic Network Embeddings for Network Evolution Analysis
Jun 24, 2019



Abstract:Network embeddings learn to represent nodes as low-dimensional vectors to preserve the proximity between nodes and communities of the network for network analysis. The temporal edges (e.g., relationships, contacts, and emails) in dynamic networks are important for network evolution analysis, but few existing methods in network embeddings can capture the dynamic information from temporal edges. In this paper, we propose a novel dynamic network embedding method to analyze evolution patterns of dynamic networks effectively. Our method uses random walk to keep the proximity between nodes and applies dynamic Bernoulli embeddings to train discrete-time network embeddings in the same vector space without alignments to preserve the temporal continuity of stable nodes. We compare our method with several state-of-the-art methods by link prediction and evolving node detection, and the experiments demonstrate that our method generally has better performance in these tasks. Our method is further verified by two real-world dynamic networks via detecting evolving nodes and visualizing their temporal trajectories in the embedded space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge