Yu-Lun Lo
A persistent homology approach to heart rate variability analysis with an application to sleep-wake classification
Aug 09, 2019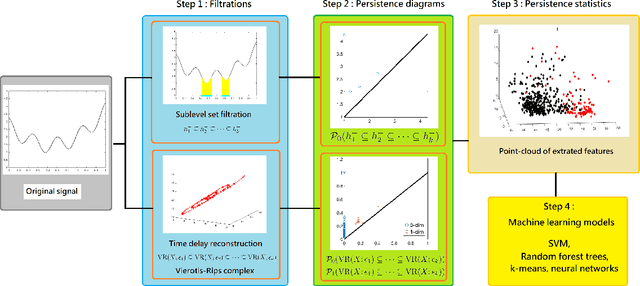
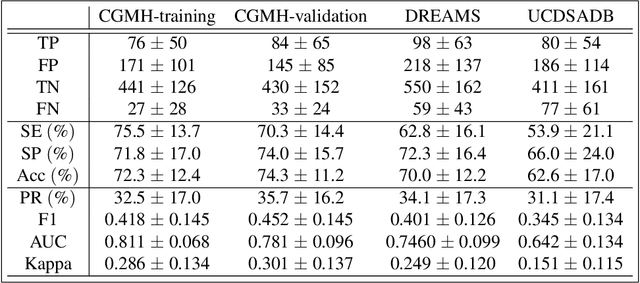
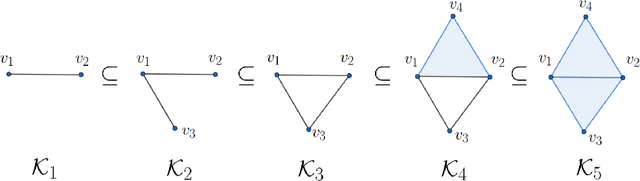
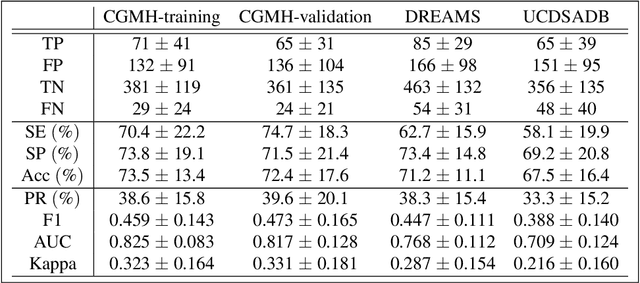
Abstract:Persistent homology (PH) is a recently developed theory in the field of algebraic topology. It is an effective and robust tool to study shapes of datasets and has been widely applied. We demonstrate a general pipeline to apply PH to study time series; particularly the heart rate variability (HRV). First, we study the shapes of time series in two different ways -- sub-level set and Taken's lag map. Second, we propose a systematic approach to summarize/vectorize persistence diagrams, a companion tool of PH. To demonstrate our proposed method, we apply these tools to the HRV analysis and the sleep-wake, REM-NREM (rapid eyeball movement and non rapid eyeball movement) and sleep-REM-NREM classification problems. The proposed algorithm is evaluated on three different datasets via the cross-database validation scheme. The performance of our approach is comparable with the state-of-the-art algorithms, and are consistent throughout these different datasets.
Calibration for massive physiological signal collection in hospital -- Sawtooth artifact in beat-to-beat pulse transit time measured from patient monitor data
Aug 27, 2018



Abstract:Objective: Calibration is one of the most important initial steps in any signal acquisition and experiment. It is however less discussed when massively collecting physiological signals in clinical setting. Here we test an off-the-shelf integrated Photoplethysmography (PPG) and electrocardiogram (ECG) monitoring device for its ability to yield a stable Pulse transit time (PTT) signal. Method: This is a retrospective clinical study using two databases: one containing 35 subjects who underwent laparoscopic cholecystectomy, another containing 22 subjects who underwent spontaneous breathing test in the intensive care unit. All data sets include recordings of PPG and ECG using a commonly deployed patient monitor. We calculated the PTT signal offline. Result: We identify a novel constant oscillatory pattern in the PTT signal and identify this pattern as the sawtooth artifact. We propose an approach based on the de-shape method to visualize, quantify and validate this sawtooth artifact. Conclusion: The PTT and ECG signals not designed for the PTT evaluation may contain unwanted artifacts. The PTT signal should be calibrated before analysis to avoid erroneous interpretation of its physiological meaning.
Sleep-wake classification via quantifying heart rate variability by convolutional neural network
Aug 01, 2018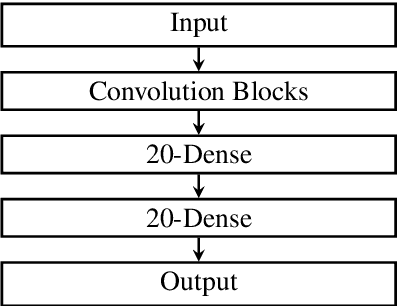
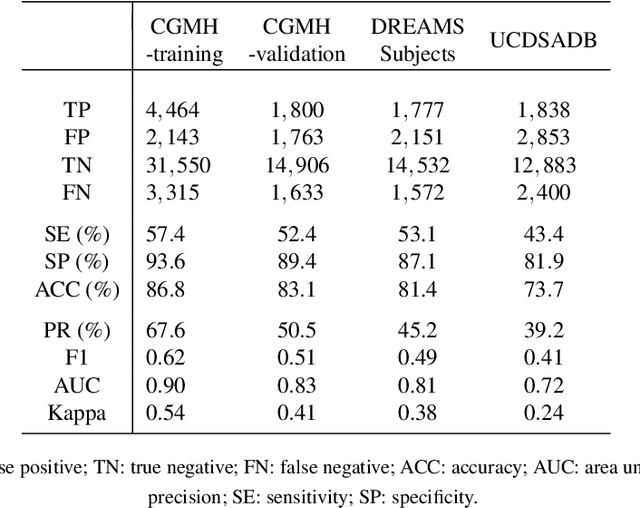
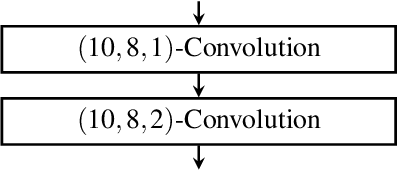
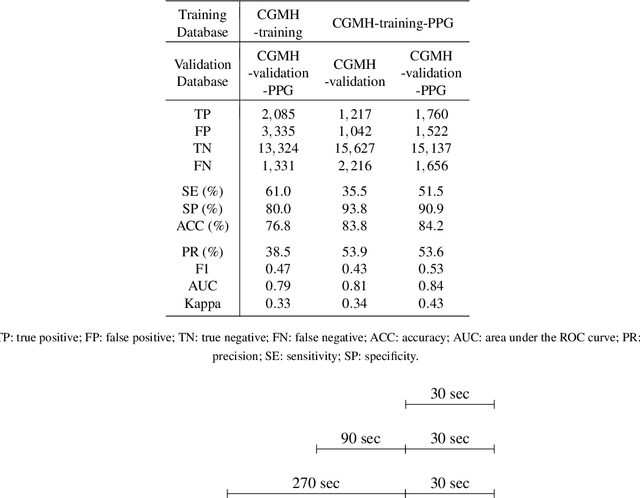
Abstract:Fluctuations in heart rate are intimately tied to changes in the physiological state of the organism. We examine and exploit this relationship by classifying a human subject's wake/sleep status using his instantaneous heart rate (IHR) series. We use a convolutional neural network (CNN) to build features from the IHR series extracted from a whole-night electrocardiogram (ECG) and predict every 30 seconds whether the subject is awake or asleep. Our training database consists of 56 normal subjects, and we consider three different databases for validation; one is private, and two are public with different races and apnea severities. On our private database of 27 subjects, our accuracy, sensitivity, specificity, and AUC values for predicting the wake stage are 83.1%, 52.4%, 89.4%, and 0.83, respectively. Validation performance is similar on our two public databases. When we use the photoplethysmography instead of the ECG to obtain the IHR series, the performance is also comparable. A robustness check is carried out to confirm the obtained performance statistics. This result advocates for an effective and scalable method for recognizing changes in physiological state using non-invasive heart rate monitoring. The CNN model adaptively quantifies IHR fluctuation as well as its location in time and is suitable for differentiating between the wake and sleep stages.
Diffusion-based nonlinear filtering for multimodal data fusion with application to sleep stage assessment
Jan 13, 2017



Abstract:The problem of information fusion from multiple data-sets acquired by multimodal sensors has drawn significant research attention over the years. In this paper, we focus on a particular problem setting consisting of a physical phenomenon or a system of interest observed by multiple sensors. We assume that all sensors measure some aspects of the system of interest with additional sensor-specific and irrelevant components. Our goal is to recover the variables relevant to the observed system and to filter out the nuisance effects of the sensor-specific variables. We propose an approach based on manifold learning, which is particularly suitable for problems with multiple modalities, since it aims to capture the intrinsic structure of the data and relies on minimal prior model knowledge. Specifically, we propose a nonlinear filtering scheme, which extracts the hidden sources of variability captured by two or more sensors, that are independent of the sensor-specific components. In addition to presenting a theoretical analysis, we demonstrate our technique on real measured data for the purpose of sleep stage assessment based on multiple, multimodal sensor measurements. We show that without prior knowledge on the different modalities and on the measured system, our method gives rise to a data-driven representation that is well correlated with the underlying sleep process and is robust to noise and sensor-specific effects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge