Yoshihiro Yamanishi
A Reinforcement Learning-Driven Transformer GAN for Molecular Generation
Mar 17, 2025Abstract:Generating molecules with desired chemical properties presents a critical challenge in fields such as chemical synthesis and drug discovery. Recent advancements in artificial intelligence (AI) and deep learning have significantly contributed to data-driven molecular generation. However, challenges persist due to the inherent sensitivity of simplified molecular input line entry system (SMILES) representations and the difficulties in applying generative adversarial networks (GANs) to discrete data. This study introduces RL-MolGAN, a novel Transformer-based discrete GAN framework designed to address these challenges. Unlike traditional Transformer architectures, RL-MolGAN utilizes a first-decoder-then-encoder structure, facilitating the generation of drug-like molecules from both $de~novo$ and scaffold-based designs. In addition, RL-MolGAN integrates reinforcement learning (RL) and Monte Carlo tree search (MCTS) techniques to enhance the stability of GAN training and optimize the chemical properties of the generated molecules. To further improve the model's performance, RL-MolWGAN, an extension of RL-MolGAN, incorporates Wasserstein distance and mini-batch discrimination, which together enhance the stability of the GAN. Experimental results on two widely used molecular datasets, QM9 and ZINC, validate the effectiveness of our models in generating high-quality molecular structures with diverse and desirable chemical properties.
Gx2Mol: De Novo Generation of Hit-like Molecules from Gene Expression Profiles via Deep Learning
Dec 27, 2024Abstract:De novo generation of hit-like molecules is a challenging task in the drug discovery process. Most methods in previous studies learn the semantics and syntax of molecular structures by analyzing molecular graphs or simplified molecular input line entry system (SMILES) strings; however, they do not take into account the drug responses of the biological systems consisting of genes and proteins. In this study we propose a deep generative model, Gx2Mol, which utilizes gene expression profiles to generate molecular structures with desirable phenotypes for arbitrary target proteins. In the algorithm, a variational autoencoder is employed as a feature extractor to learn the latent feature distribution of the gene expression profiles. Then, a long short-term memory is leveraged as the chemical generator to produce syntactically valid SMILES strings that satisfy the feature conditions of the gene expression profile extracted by the feature extractor. Experimental results and case studies demonstrate that the proposed Gx2Mol model can produce new molecules with potential bioactivities and drug-like properties.
Molecular Generative Adversarial Network with Multi-Property Optimization
Mar 29, 2024



Abstract:Deep generative models, such as generative adversarial networks (GANs), have been employed for $de~novo$ molecular generation in drug discovery. Most prior studies have utilized reinforcement learning (RL) algorithms, particularly Monte Carlo tree search (MCTS), to handle the discrete nature of molecular representations in GANs. However, due to the inherent instability in training GANs and RL models, along with the high computational cost associated with MCTS sampling, MCTS RL-based GANs struggle to scale to large chemical databases. To tackle these challenges, this study introduces a novel GAN based on actor-critic RL with instant and global rewards, called InstGAN, to generate molecules at the token-level with multi-property optimization. Furthermore, maximized information entropy is leveraged to alleviate the mode collapse. The experimental results demonstrate that InstGAN outperforms other baselines, achieves comparable performance to state-of-the-art models, and efficiently generates molecules with multi-property optimization. The source code will be released upon acceptance of the paper.
Scalable Alignment Kernels via Space-Efficient Feature Maps
Mar 28, 2018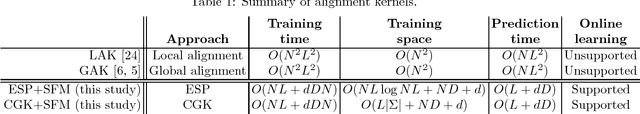

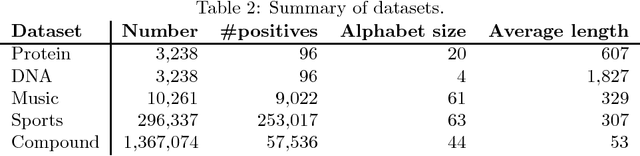
Abstract:String kernels are attractive data analysis tools for analyzing string data. Among them, alignment kernels are known for their high prediction accuracies in string classifications when tested in combination with SVMs in various applications. However, alignment kernels have a crucial drawback in that they scale poorly due to their quadratic computation complexity in the number of input strings, which limits large-scale applications in practice. We present the first approximation named ESP+SFM for alignment kernels by leveraging a metric embedding named edit-sensitive parsing (ESP) and space-efficient feature maps (SFM) for random Fourier features (RFF) for large-scale string analyses. Input strings are projected into vectors of RFF by leveraging ESP and SFM. Then, SVMs are trained on the projected vectors, which enables to significantly improve the scalability of alignment kernels while preserving their prediction accuracies. We experimentally test ESP+ SFM on its ability to learn SVMs for large-scale string classifications with various massive string data, and we demonstrate the superior performance of ESP+SFM with respect to prediction accuracy, scalability and computation efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge