Yong-Yeon Jo
ALFRED: Ask a Large-language model For Reliable ECG Diagnosis
Apr 30, 2025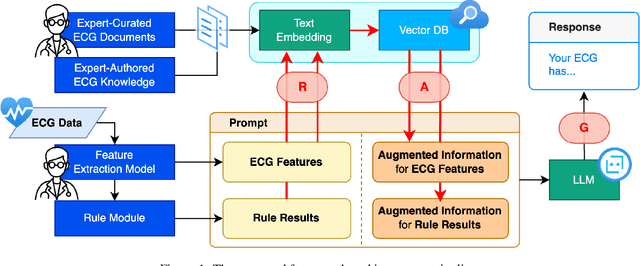
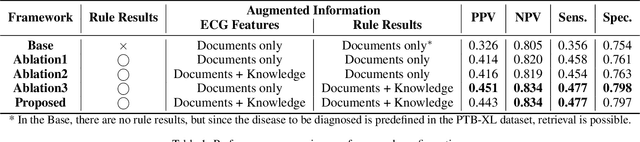
Abstract:Leveraging Large Language Models (LLMs) with Retrieval-Augmented Generation (RAG) for analyzing medical data, particularly Electrocardiogram (ECG), offers high accuracy and convenience. However, generating reliable, evidence-based results in specialized fields like healthcare remains a challenge, as RAG alone may not suffice. We propose a Zero-shot ECG diagnosis framework based on RAG for ECG analysis that incorporates expert-curated knowledge to enhance diagnostic accuracy and explainability. Evaluation on the PTB-XL dataset demonstrates the framework's effectiveness, highlighting the value of structured domain expertise in automated ECG interpretation. Our framework is designed to support comprehensive ECG analysis, addressing diverse diagnostic needs with potential applications beyond the tested dataset.
New Test-Time Scenario for Biosignal: Concept and Its Approach
Nov 26, 2024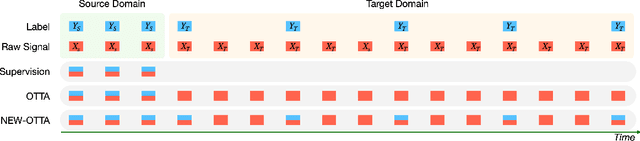
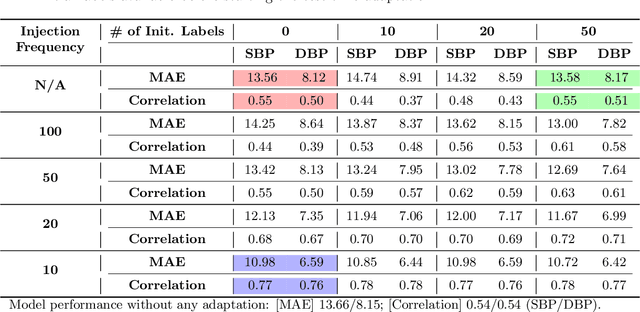
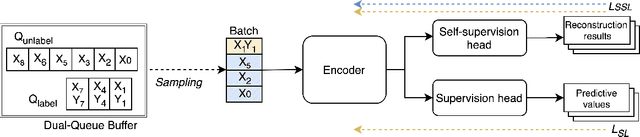
Abstract:Online Test-Time Adaptation (OTTA) enhances model robustness by updating pre-trained models with unlabeled data during testing. In healthcare, OTTA is vital for real-time tasks like predicting blood pressure from biosignals, which demand continuous adaptation. We introduce a new test-time scenario with streams of unlabeled samples and occasional labeled samples. Our framework combines supervised and self-supervised learning, employing a dual-queue buffer and weighted batch sampling to balance data types. Experiments show improved accuracy and adaptability under real-world conditions.
TADA: Temporal Adversarial Data Augmentation for Time Series Data
Jul 21, 2024Abstract:Domain generalization involves training machine learning models to perform robustly on unseen samples from out-of-distribution datasets. Adversarial Data Augmentation (ADA) is a commonly used approach that enhances model adaptability by incorporating synthetic samples, designed to simulate potential unseen samples. While ADA effectively addresses amplitude-related distribution shifts, it falls short in managing temporal shifts, which are essential for time series data. To address this limitation, we propose the Temporal Adversarial Data Augmentation for time teries Data (TADA), which incorporates a time warping technique specifically targeting temporal shifts. Recognizing the challenge of non-differentiability in traditional time warping, we make it differentiable by leveraging phase shifts in the frequency domain. Our evaluations across diverse domains demonstrate that TADA significantly outperforms existing ADA variants, enhancing model performance across time series datasets with varied distributions.
Foundation Models for Electrocardiograms
Jun 26, 2024



Abstract:Foundation models, enhanced by self-supervised learning (SSL) techniques, represent a cutting-edge frontier in biomedical signal analysis, particularly for electrocardiograms (ECGs), crucial for cardiac health monitoring and diagnosis. This study conducts a comprehensive analysis of foundation models for ECGs by employing and refining innovative SSL methodologies - namely, generative and contrastive learning - on a vast dataset of over 1.1 million ECG samples. By customizing these methods to align with the intricate characteristics of ECG signals, our research has successfully developed foundation models that significantly elevate the precision and reliability of cardiac diagnostics. These models are adept at representing the complex, subtle nuances of ECG data, thus markedly enhancing diagnostic capabilities. The results underscore the substantial potential of SSL-enhanced foundation models in clinical settings and pave the way for extensive future investigations into their scalable applications across a broader spectrum of medical diagnostics. This work sets a benchmark in the ECG field, demonstrating the profound impact of tailored, data-driven model training on the efficacy and accuracy of medical diagnostics.
Optimizing Neural Network Scale for ECG Classification
Aug 24, 2023Abstract:We study scaling convolutional neural networks (CNNs), specifically targeting Residual neural networks (ResNet), for analyzing electrocardiograms (ECGs). Although ECG signals are time-series data, CNN-based models have been shown to outperform other neural networks with different architectures in ECG analysis. However, most previous studies in ECG analysis have overlooked the importance of network scaling optimization, which significantly improves performance. We explored and demonstrated an efficient approach to scale ResNet by examining the effects of crucial parameters, including layer depth, the number of channels, and the convolution kernel size. Through extensive experiments, we found that a shallower network, a larger number of channels, and smaller kernel sizes result in better performance for ECG classifications. The optimal network scale might differ depending on the target task, but our findings provide insight into obtaining more efficient and accurate models with fewer computing resources or less time. In practice, we demonstrate that a narrower search space based on our findings leads to higher performance.
ECGT2T: Electrocardiogram synthesis from Two asynchronous leads to Ten leads
Feb 28, 2021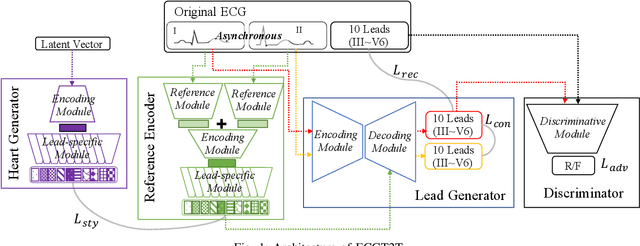
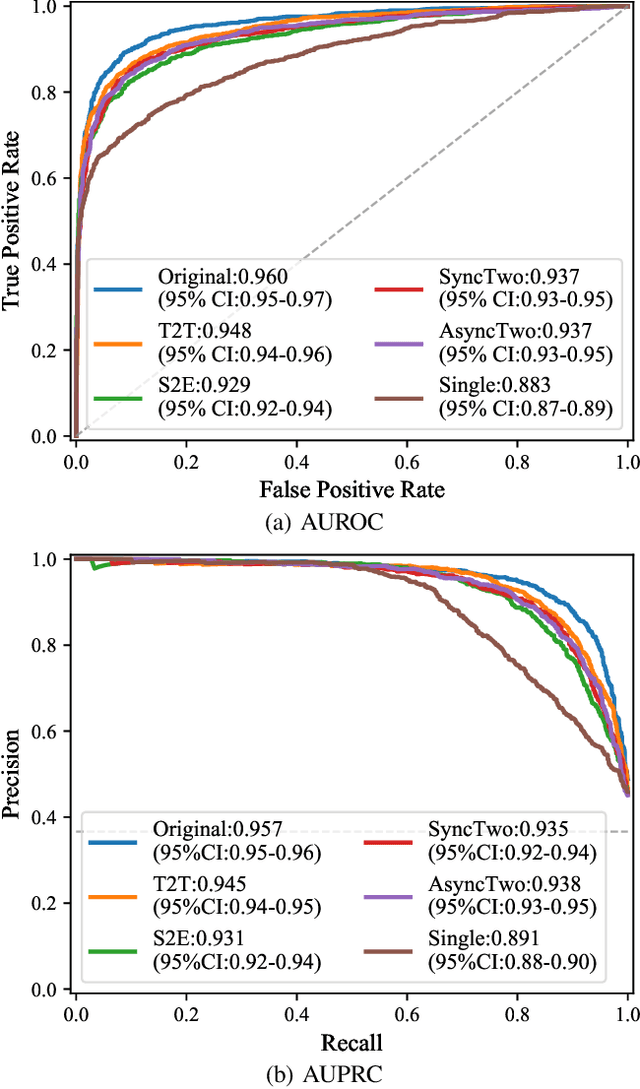
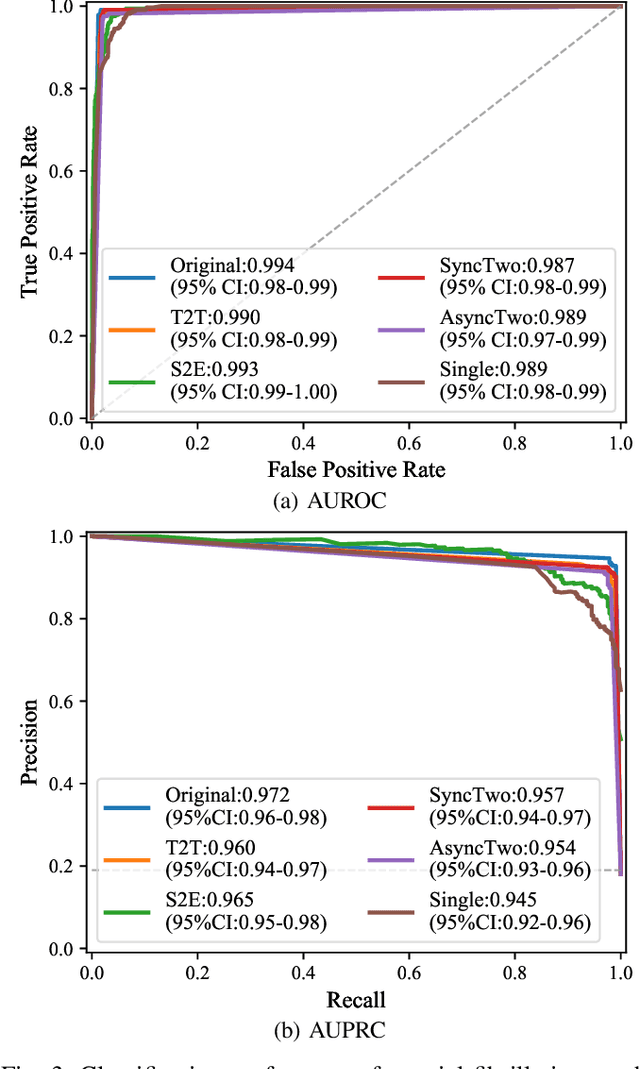
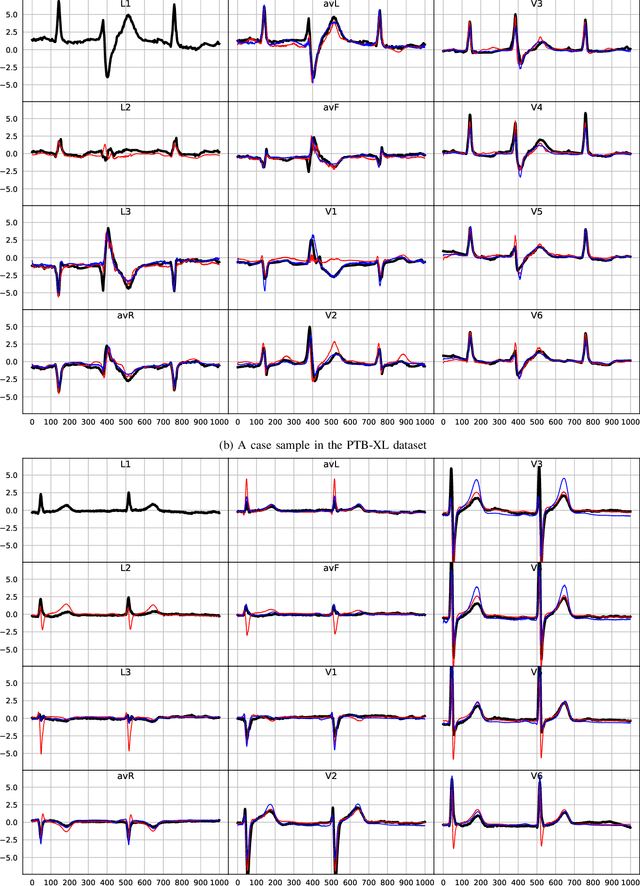
Abstract:The electrocardiogram (ECG) records electrical signals in a non-invasive way to observe the condition of the heart. It consists of 12 leads that look at the heart from different directions. Recently, various wearable devices have enabled immediate access to the ECG without the use of wieldy equipment. However, they only provide ECGs with one or two leads. This results in an inaccurate diagnosis of cardiac disease. We propose a deep generative model for ECG synthesis from two asynchronous leads to ten leads (ECGT2T). It first represents a heart condition referring to two leads, and then generates ten leads based on the represented heart condition. Both the rhythm and amplitude of leads generated by ECGT2T resemble those of the original ones, while the technique removes noise and the baseline wander appearing in the original leads. As a data augmentation method, ECGT2T improves the classification performance of models compared with models using ECGs with a couple of leads.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge