Joon-Myoung Kwon
Optimizing Neural Network Scale for ECG Classification
Aug 24, 2023Abstract:We study scaling convolutional neural networks (CNNs), specifically targeting Residual neural networks (ResNet), for analyzing electrocardiograms (ECGs). Although ECG signals are time-series data, CNN-based models have been shown to outperform other neural networks with different architectures in ECG analysis. However, most previous studies in ECG analysis have overlooked the importance of network scaling optimization, which significantly improves performance. We explored and demonstrated an efficient approach to scale ResNet by examining the effects of crucial parameters, including layer depth, the number of channels, and the convolution kernel size. Through extensive experiments, we found that a shallower network, a larger number of channels, and smaller kernel sizes result in better performance for ECG classifications. The optimal network scale might differ depending on the target task, but our findings provide insight into obtaining more efficient and accurate models with fewer computing resources or less time. In practice, we demonstrate that a narrower search space based on our findings leads to higher performance.
ECGT2T: Electrocardiogram synthesis from Two asynchronous leads to Ten leads
Feb 28, 2021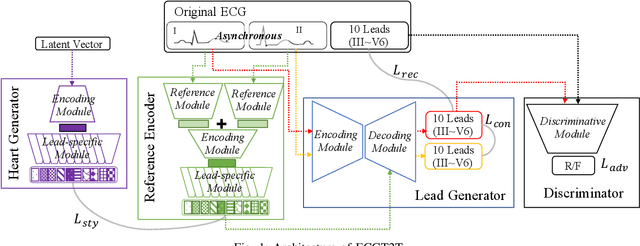
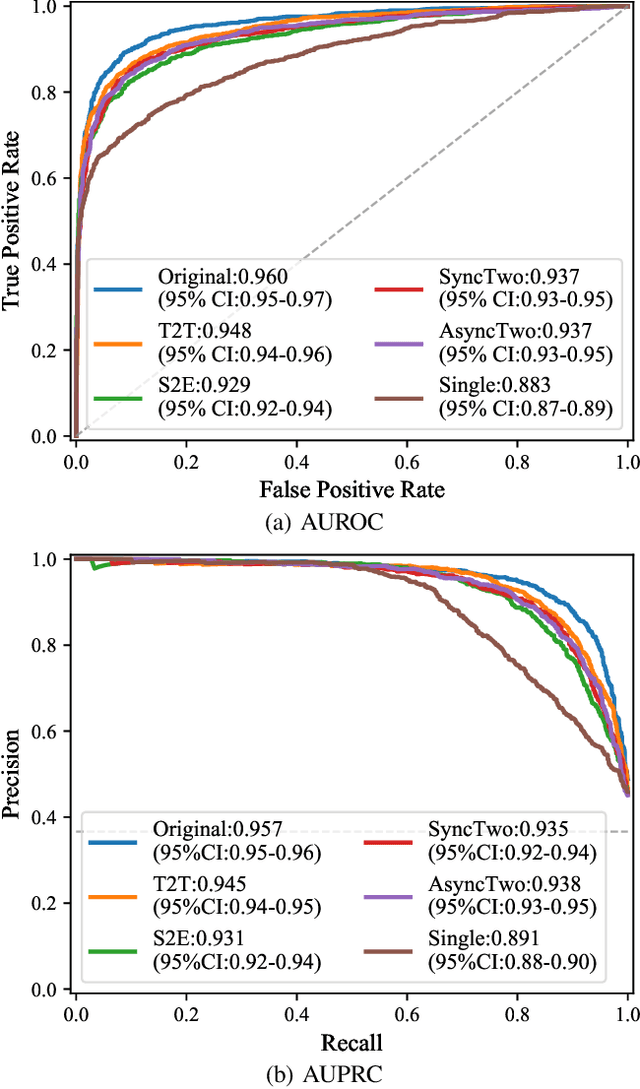
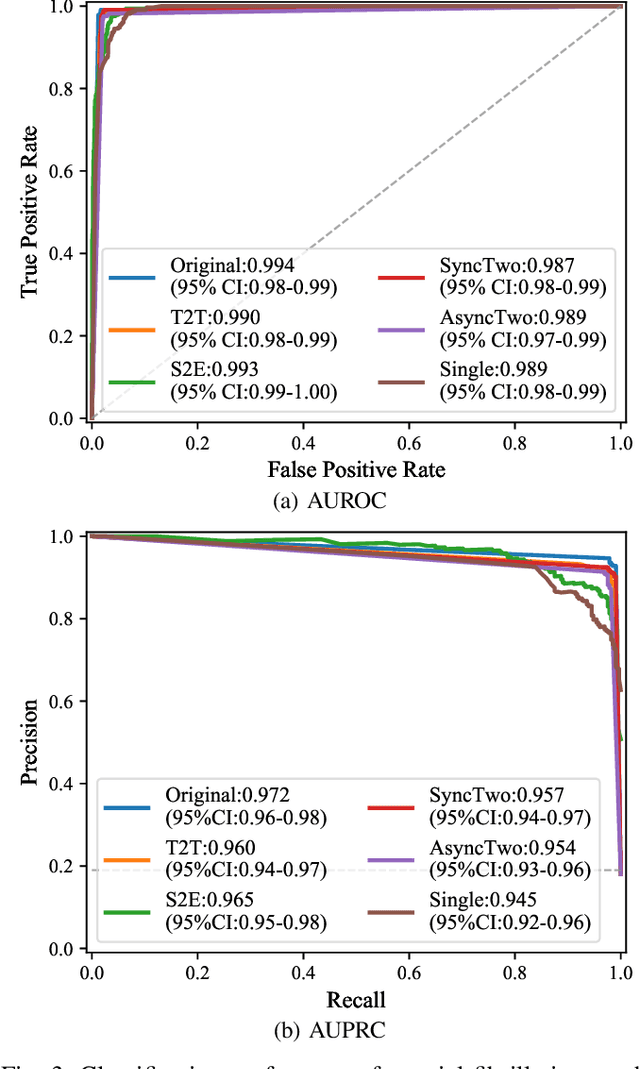
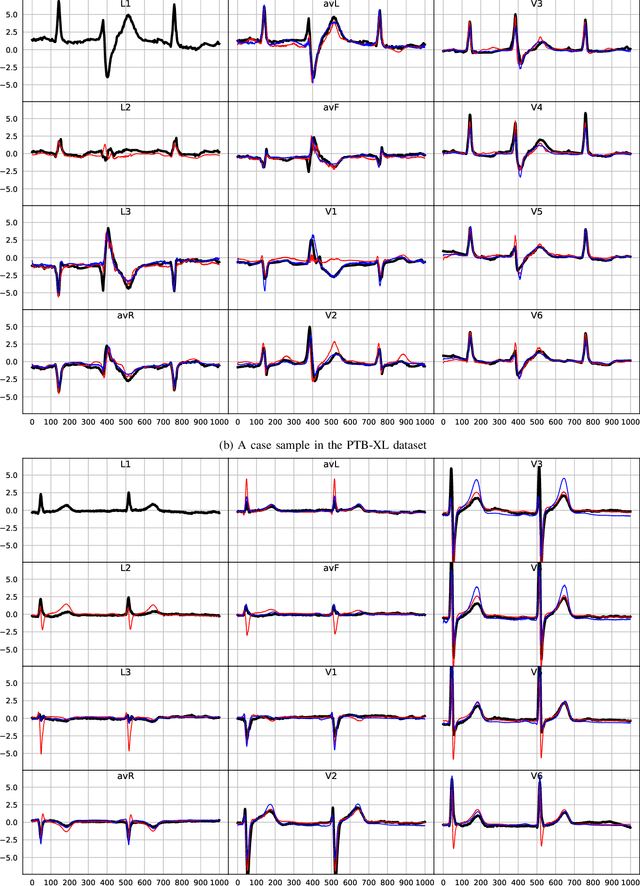
Abstract:The electrocardiogram (ECG) records electrical signals in a non-invasive way to observe the condition of the heart. It consists of 12 leads that look at the heart from different directions. Recently, various wearable devices have enabled immediate access to the ECG without the use of wieldy equipment. However, they only provide ECGs with one or two leads. This results in an inaccurate diagnosis of cardiac disease. We propose a deep generative model for ECG synthesis from two asynchronous leads to ten leads (ECGT2T). It first represents a heart condition referring to two leads, and then generates ten leads based on the represented heart condition. Both the rhythm and amplitude of leads generated by ECGT2T resemble those of the original ones, while the technique removes noise and the baseline wander appearing in the original leads. As a data augmentation method, ECGT2T improves the classification performance of models compared with models using ECGs with a couple of leads.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge