Byeong Tak Lee
New Test-Time Scenario for Biosignal: Concept and Its Approach
Nov 26, 2024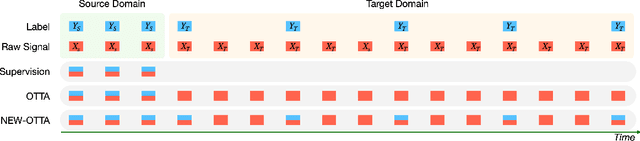
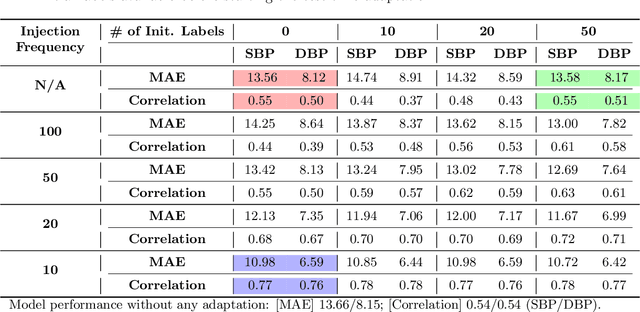
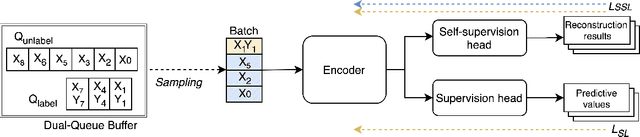
Abstract:Online Test-Time Adaptation (OTTA) enhances model robustness by updating pre-trained models with unlabeled data during testing. In healthcare, OTTA is vital for real-time tasks like predicting blood pressure from biosignals, which demand continuous adaptation. We introduce a new test-time scenario with streams of unlabeled samples and occasional labeled samples. Our framework combines supervised and self-supervised learning, employing a dual-queue buffer and weighted batch sampling to balance data types. Experiments show improved accuracy and adaptability under real-world conditions.
TADA: Temporal Adversarial Data Augmentation for Time Series Data
Jul 21, 2024Abstract:Domain generalization involves training machine learning models to perform robustly on unseen samples from out-of-distribution datasets. Adversarial Data Augmentation (ADA) is a commonly used approach that enhances model adaptability by incorporating synthetic samples, designed to simulate potential unseen samples. While ADA effectively addresses amplitude-related distribution shifts, it falls short in managing temporal shifts, which are essential for time series data. To address this limitation, we propose the Temporal Adversarial Data Augmentation for time teries Data (TADA), which incorporates a time warping technique specifically targeting temporal shifts. Recognizing the challenge of non-differentiability in traditional time warping, we make it differentiable by leveraging phase shifts in the frequency domain. Our evaluations across diverse domains demonstrate that TADA significantly outperforms existing ADA variants, enhancing model performance across time series datasets with varied distributions.
Foundation Models for Electrocardiograms
Jun 26, 2024



Abstract:Foundation models, enhanced by self-supervised learning (SSL) techniques, represent a cutting-edge frontier in biomedical signal analysis, particularly for electrocardiograms (ECGs), crucial for cardiac health monitoring and diagnosis. This study conducts a comprehensive analysis of foundation models for ECGs by employing and refining innovative SSL methodologies - namely, generative and contrastive learning - on a vast dataset of over 1.1 million ECG samples. By customizing these methods to align with the intricate characteristics of ECG signals, our research has successfully developed foundation models that significantly elevate the precision and reliability of cardiac diagnostics. These models are adept at representing the complex, subtle nuances of ECG data, thus markedly enhancing diagnostic capabilities. The results underscore the substantial potential of SSL-enhanced foundation models in clinical settings and pave the way for extensive future investigations into their scalable applications across a broader spectrum of medical diagnostics. This work sets a benchmark in the ECG field, demonstrating the profound impact of tailored, data-driven model training on the efficacy and accuracy of medical diagnostics.
Self Attention with Temporal Prior: Can We Learn More from Arrow of Time?
Oct 29, 2023Abstract:Many of diverse phenomena in nature often inherently encode both short and long term temporal dependencies, short term dependencies especially resulting from the direction of flow of time. In this respect, we discovered experimental evidences suggesting that {\it interrelations} of these events are higher for closer time stamps. However, to be able for attention based models to learn these regularities in short term dependencies, it requires large amounts of data which are often infeasible. This is due to the reason that, while they are good at learning piece wised temporal dependencies, attention based models lack structures that encode biases in time series. As a resolution, we propose a simple and efficient method that enables attention layers to better encode short term temporal bias of these data sets by applying learnable, adaptive kernels directly to the attention matrices. For the experiments, we chose various prediction tasks using Electronic Health Records (EHR) data sets since they are great examples that have underlying long and short term temporal dependencies. The results of our experiments show exceptional classification results compared to best performing models on most of the task and data sets.
Optimizing Neural Network Scale for ECG Classification
Aug 24, 2023Abstract:We study scaling convolutional neural networks (CNNs), specifically targeting Residual neural networks (ResNet), for analyzing electrocardiograms (ECGs). Although ECG signals are time-series data, CNN-based models have been shown to outperform other neural networks with different architectures in ECG analysis. However, most previous studies in ECG analysis have overlooked the importance of network scaling optimization, which significantly improves performance. We explored and demonstrated an efficient approach to scale ResNet by examining the effects of crucial parameters, including layer depth, the number of channels, and the convolution kernel size. Through extensive experiments, we found that a shallower network, a larger number of channels, and smaller kernel sizes result in better performance for ECG classifications. The optimal network scale might differ depending on the target task, but our findings provide insight into obtaining more efficient and accurate models with fewer computing resources or less time. In practice, we demonstrate that a narrower search space based on our findings leads to higher performance.
Graph Structure Based Data Augmentation Method
May 29, 2022
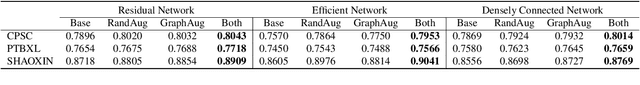
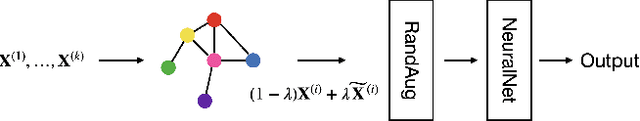

Abstract:In this paper, we propose a novel graph-based data augmentation method that can generally be applied to medical waveform data with graph structures. In the process of recording medical waveform data, such as electrocardiogram (ECG) or electroencephalogram (EEG), angular perturbations between the measurement leads exist due to discrepancies in lead positions. The data samples with large angular perturbations often cause inaccuracy in algorithmic prediction tasks. We design a graph-based data augmentation technique that exploits the inherent graph structures within the medical waveform data to improve both performance and robustness. In addition, we show that the performance gain from graph augmentation results from robustness by testing against adversarial attacks. Since the bases of performance gain are orthogonal, the graph augmentation can be used in conjunction with existing data augmentation techniques to further improve the final performance. We believe that our graph augmentation method opens up new possibilities to explore in data augmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge