Yishen Liu
Hengqin-RA-v1: Advanced Large Language Model for Diagnosis and Treatment of Rheumatoid Arthritis with Dataset based Traditional Chinese Medicine
Jan 05, 2025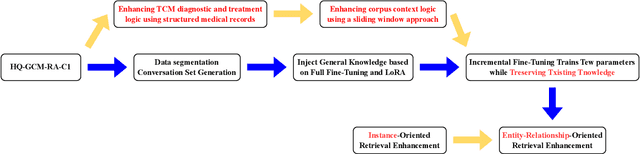
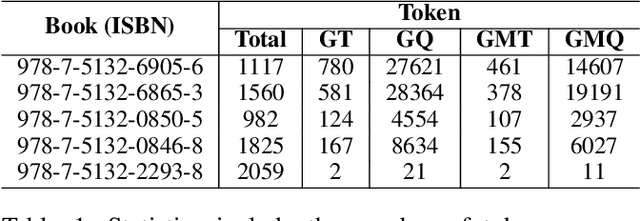
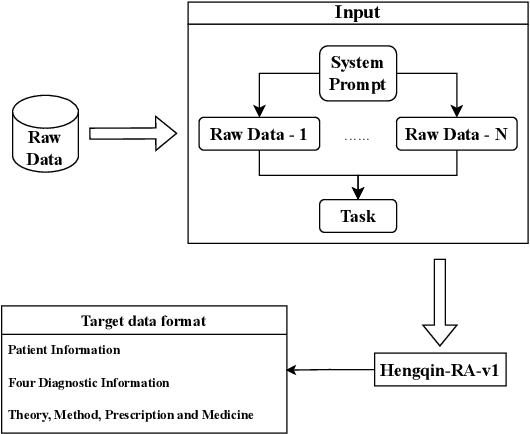
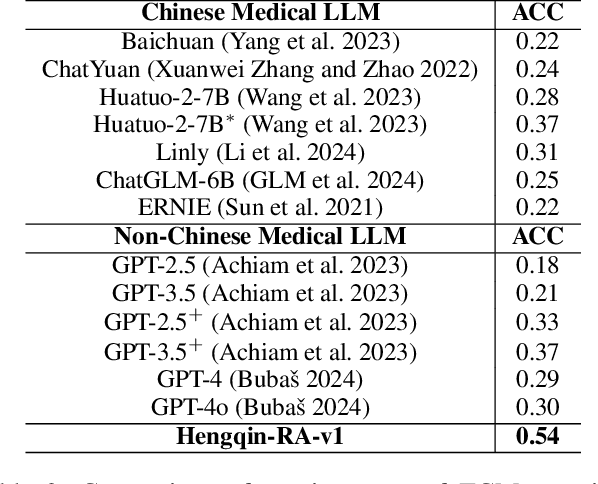
Abstract:Large language models (LLMs) primarily trained on English texts, often face biases and inaccuracies in Chinese contexts. Their limitations are pronounced in fields like Traditional Chinese Medicine (TCM), where cultural and clinical subtleties are vital, further hindered by a lack of domain-specific data, such as rheumatoid arthritis (RA). To address these issues, this paper introduces Hengqin-RA-v1, the first large language model specifically tailored for TCM with a focus on diagnosing and treating RA. We also present HQ-GCM-RA-C1, a comprehensive RA-specific dataset curated from ancient Chinese medical literature, classical texts, and modern clinical studies. This dataset empowers Hengqin-RA-v1 to deliver accurate and culturally informed responses, effectively bridging the gaps left by general-purpose models. Extensive experiments demonstrate that Hengqin-RA-v1 outperforms state-of-the-art models, even surpassing the diagnostic accuracy of TCM practitioners in certain cases.
Latent Relationship Mining of Glaucoma Biomarkers: a TRI-LSTM based Deep Learning
Aug 28, 2024



Abstract:In recently years, a significant amount of research has been conducted on applying deep learning methods for glaucoma classification and detection. However, the explainability of those established machine learning models remains a big concern. In this research, in contrast, we learn from cognitive science concept and study how ophthalmologists judge glaucoma detection. Simulating experts' efforts, we propose a hierarchical decision making system, centered around a holistic set of carefully designed biomarker-oriented machine learning models. While biomarkers represent the key indicators of how ophthalmologists identify glaucoma, they usually exhibit latent inter-relations. We thus construct a time series model, named TRI-LSTM, capable of calculating and uncovering potential and latent relationships among various biomarkers of glaucoma. Our model is among the first efforts to explore the intrinsic connections among glaucoma biomarkers. We monitor temporal relationships in patients' disease states over time and to capture and retain the progression of disease-relevant clinical information from prior visits, thereby enriching biomarker's potential relationships. Extensive experiments over real-world dataset have demonstrated the effectiveness of the proposed model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge