Yingtao Liu
Digital Twins in Additive Manufacturing: A Systematic Review
Sep 02, 2024Abstract:Digital Twins (DTs) are becoming popular in Additive Manufacturing (AM) due to their ability to create virtual replicas of physical components of AM machines, which helps in real-time production monitoring. Advanced techniques such as Machine Learning (ML), Augmented Reality (AR), and simulation-based models play key roles in developing intelligent and adaptable DTs in manufacturing processes. However, questions remain regarding scalability, the integration of high-quality data, and the computational power required for real-time applications in developing DTs. Understanding the current state of DTs in AM is essential to address these challenges and fully utilize their potential in advancing AM processes. Considering this opportunity, this work aims to provide a comprehensive overview of DTs in AM by addressing the following four research questions: (1) What are the key types of DTs used in AM and their specific applications? (2) What are the recent developments and implementations of DTs? (3) How are DTs employed in process improvement and hybrid manufacturing? (4) How are DTs integrated with Industry 4.0 technologies? By discussing current applications and techniques, we aim to offer a better understanding and potential future research directions for researchers and practitioners in AM and DTs.
AlphaFold Accelerates Artificial Intelligence Powered Drug Discovery: Efficient Discovery of a Novel Cyclin-dependent Kinase 20 (CDK20) Small Molecule Inhibitor
Jan 21, 2022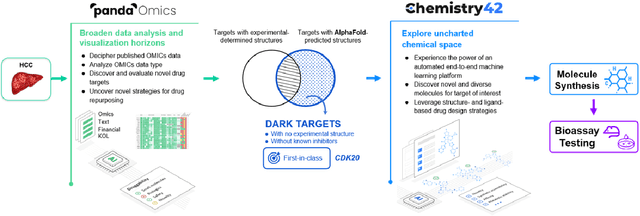
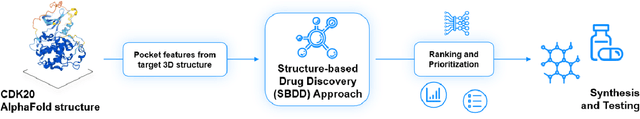
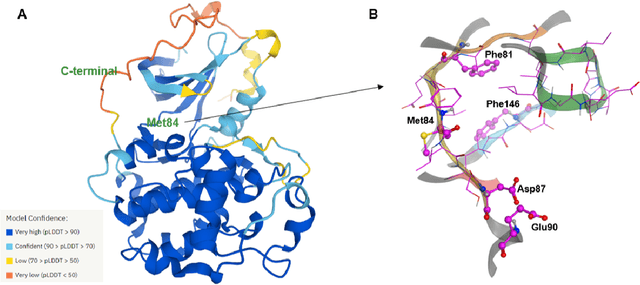
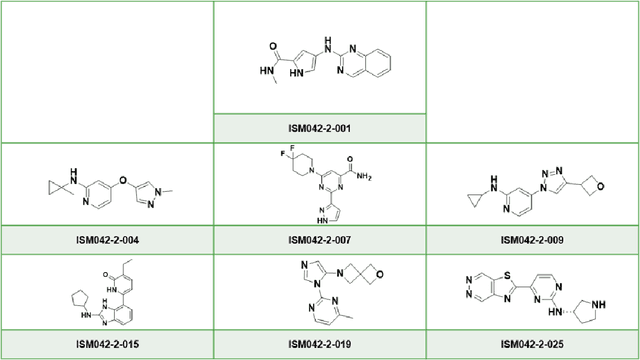
Abstract:The AlphaFold computer program predicted protein structures for the whole human genome, which has been considered as a remarkable breakthrough both in artificial intelligence (AI) application and structural biology. Despite the varying confidence level, these predicted structures still could significantly contribute to the structure-based drug design of novel targets, especially the ones with no or limited structural information. In this work, we successfully applied AlphaFold in our end-to-end AI-powered drug discovery engines constituted of a biocomputational platform PandaOmics and a generative chemistry platform Chemistry42, to identify a first-in-class hit molecule of a novel target without an experimental structure starting from target selection towards hit identification in a cost- and time-efficient manner. PandaOmics provided the targets of interest and Chemistry42 generated the molecules based on the AlphaFold predicted structure, and the selected molecules were synthesized and tested in biological assays. Through this approach, we identified a small molecule hit compound for CDK20 with a Kd value of 8.9 +/- 1.6 uM (n = 4) within 30 days from target selection and after only synthesizing 7 compounds. To the best of our knowledge, this is the first reported small molecule targeting CDK20 and more importantly, this work is the first demonstration of AlphaFold application in the hit identification process in early drug discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge