Yasuhiko Tachibana
Applied MRI Research, Department of Molecular imaging and Theranostics, National Institute of Radiological Sciences, QST, Department of Radiology, Juntendo University School of Medicine
A neural network model that learns differences in diagnosis strategies among radiologists has an improved area under the curve for aneurysm status classification in magnetic resonance angiography image series
Feb 03, 2020



Abstract:Purpose: To construct a neural network model that can learn the different diagnosing strategies of radiologists to better classify aneurysm status in magnetic resonance angiography images. Materials and methods: This retrospective study included 3423 time-of-flight brain magnetic resonance angiography image series (subjects: male 1843 [mean age, 50.2 +/- 11.7 years], female 1580 [50.8 +/- 11.3 years]) recorded from November 2017 through January 2019. The image series were read independently for aneurysm status by one of four board-certified radiologists, who were assisted by an established deep learning-based computer-assisted diagnosis (CAD) system. The constructed neural networks were trained to classify the aneurysm status of zero to five aneurysm-suspicious areas suggested by the CAD system for each image series, and any additional aneurysm areas added by the radiologists, and this classification was compared with the judgment of the annotating radiologist. Image series were randomly allocated to training and testing data in an 8:2 ratio. The accuracy of the classification was compared by receiver operating characteristic analysis between the control model that accepted only image data as input and the proposed model that additionally accepted the information of who the annotating radiologist was. The DeLong test was used to compare areas under the curves (P < 0.05 was considered significant). Results: The area under the curve was larger in the proposed model (0.845) than in the control model (0.793), and the difference was significant (P < 0.0001). Conclusion: The proposed model improved classification accuracy by learning the diagnosis strategies of individual annotating radiologists.
The utility of a convolutional neural network for generating a myelin volume index map from rapid simultaneous relaxometry imaging
Apr 24, 2019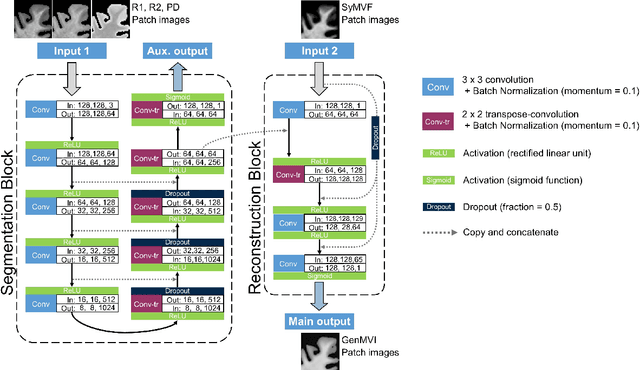
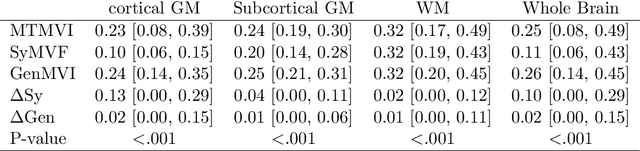
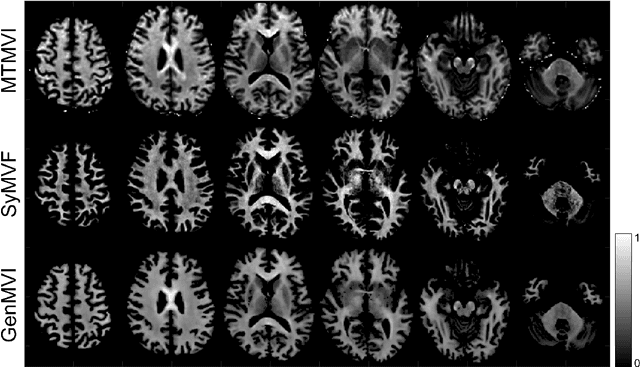
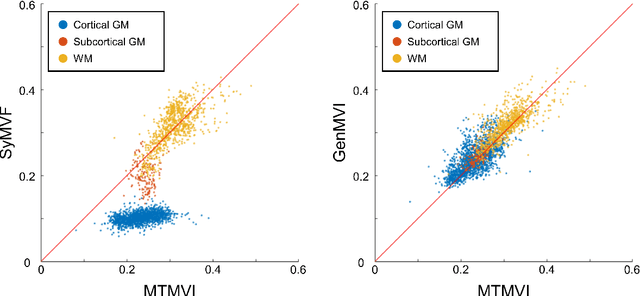
Abstract:Background and Purpose: A current algorithm to obtain a synthetic myelin volume fraction map (SyMVF) from rapid simultaneous relaxometry imaging (RSRI) has a potential problem, that it does not incorporate information from surrounding pixels. The purpose of this study was to develop a method that utilizes a convolutional neural network (CNN) to overcome this problem. Methods: RSRI and magnetization transfer images from 20 healthy volunteers were included. A CNN was trained to reconstruct RSRI-related metric maps into a myelin volume-related index (generated myelin volume index: GenMVI) map using the myelin volume index map calculated from magnetization transfer images (MTMVI) as reference. The SyMVF and GenMVI maps were statistically compared by testing how well they correlated with the MTMVI map. The correlations were evaluated based on: (i) averaged values obtained from 164 atlas-based ROIs, and (ii) pixel-based comparison for ROIs defined in four different tissue types (cortical and subcortical gray matter, white matter, and whole brain). Results: For atlas-based ROIs, the overall correlation with the MTMVI map was higher for the GenMVI map than for the SyMVF map. In the pixel-based comparison, correlation with the MTMVI map was stronger for the GenMVI map than for the SyMVF map, and the difference in the distribution for the volunteers was significant (Wilcoxon sign-rank test, P<.001) in all tissue types. Conclusion: The proposed method is useful, as it can incorporate more specific information about local tissue properties than the existing method.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018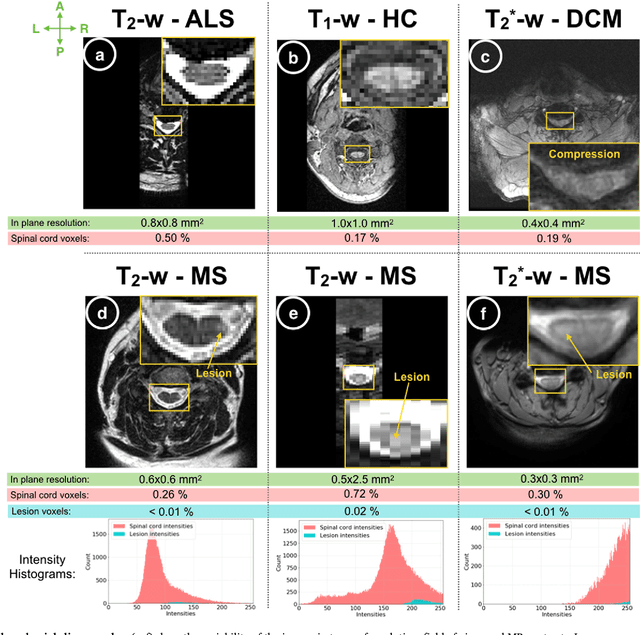
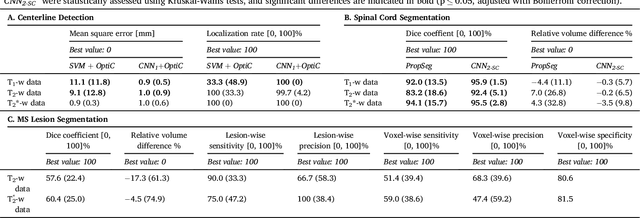
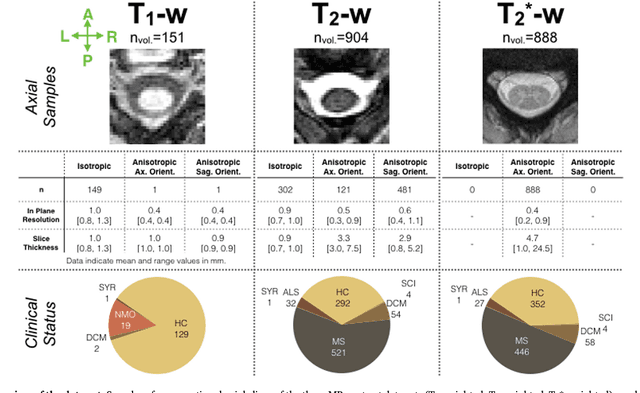
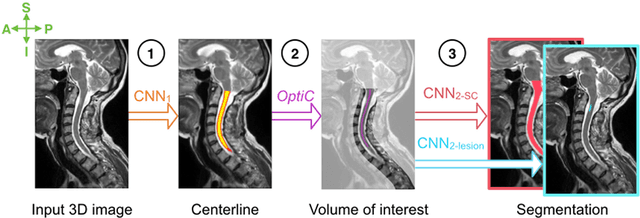
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge