Yash-yee Logan
OLIVES Dataset: Ophthalmic Labels for Investigating Visual Eye Semantics
Sep 22, 2022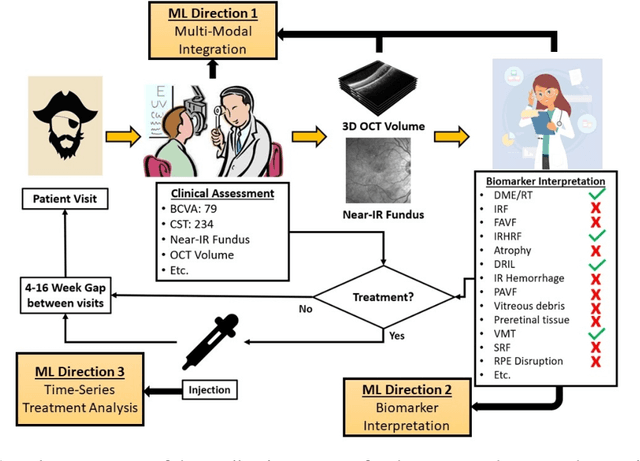
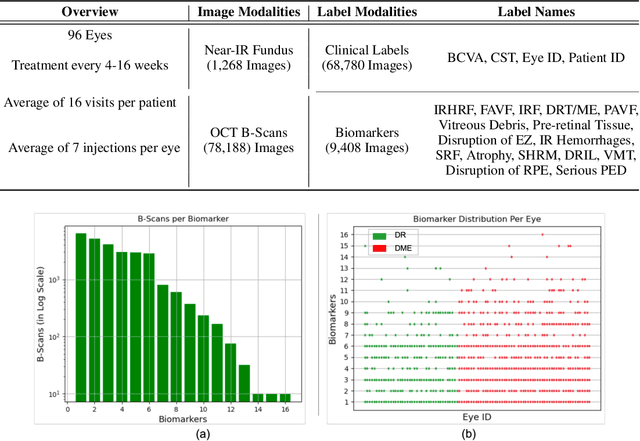
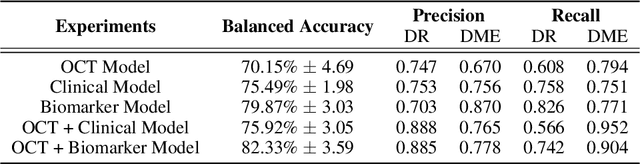
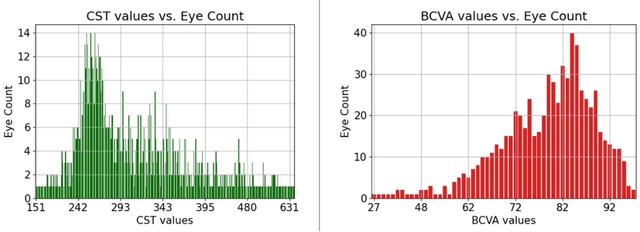
Abstract:Clinical diagnosis of the eye is performed over multifarious data modalities including scalar clinical labels, vectorized biomarkers, two-dimensional fundus images, and three-dimensional Optical Coherence Tomography (OCT) scans. Clinical practitioners use all available data modalities for diagnosing and treating eye diseases like Diabetic Retinopathy (DR) or Diabetic Macular Edema (DME). Enabling usage of machine learning algorithms within the ophthalmic medical domain requires research into the relationships and interactions between all relevant data over a treatment period. Existing datasets are limited in that they neither provide data nor consider the explicit relationship modeling between the data modalities. In this paper, we introduce the Ophthalmic Labels for Investigating Visual Eye Semantics (OLIVES) dataset that addresses the above limitation. This is the first OCT and near-IR fundus dataset that includes clinical labels, biomarker labels, disease labels, and time-series patient treatment information from associated clinical trials. The dataset consists of 1268 near-IR fundus images each with at least 49 OCT scans, and 16 biomarkers, along with 4 clinical labels and a disease diagnosis of DR or DME. In total, there are 96 eyes' data averaged over a period of at least two years with each eye treated for an average of 66 weeks and 7 injections. We benchmark the utility of OLIVES dataset for ophthalmic data as well as provide benchmarks and concrete research directions for core and emerging machine learning paradigms within medical image analysis.
Patient Aware Active Learning for Fine-Grained OCT Classification
Jun 27, 2022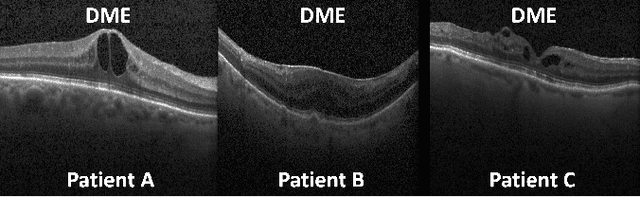

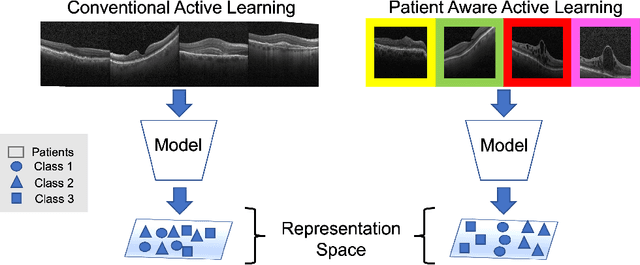
Abstract:This paper considers making active learning more sensible from a medical perspective. In practice, a disease manifests itself in different forms across patient cohorts. Existing frameworks have primarily used mathematical constructs to engineer uncertainty or diversity-based methods for selecting the most informative samples. However, such algorithms do not present themselves naturally as usable by the medical community and healthcare providers. Thus, their deployment in clinical settings is very limited, if any. For this purpose, we propose a framework that incorporates clinical insights into the sample selection process of active learning that can be incorporated with existing algorithms. Our medically interpretable active learning framework captures diverse disease manifestations from patients to improve generalization performance of OCT classification. After comprehensive experiments, we report that incorporating patient insights within the active learning framework yields performance that matches or surpasses five commonly used paradigms on two architectures with a dataset having imbalanced patient distributions. Also, the framework integrates within existing medical practices and thus can be used by healthcare providers.
DECAL: DEployable Clinical Active Learning
Jun 27, 2022



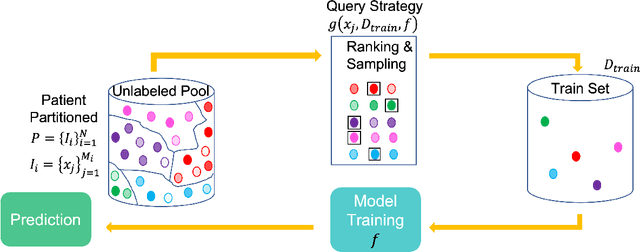
Abstract:Conventional machine learning systems that operate on natural images assume the presence of attributes within the images that lead to some decision. However, decisions in medical domain are a resultant of attributes within medical diagnostic scans and electronic medical records (EMR). Hence, active learning techniques that are developed for natural images are insufficient for handling medical data. We focus on reducing this insufficiency by designing a deployable clinical active learning (DECAL) framework within a bi-modal interface so as to add practicality to the paradigm. Our approach is a "plug-in" method that makes natural image based active learning algorithms generalize better and faster. We find that on two medical datasets on three architectures and five learning strategies, DECAL increases generalization across 20 rounds by approximately 4.81%. DECAL leads to a 5.59% and 7.02% increase in average accuracy as an initialization strategy for optical coherence tomography (OCT) and X-Ray respectively. Our active learning results were achieved using 3000 (5%) and 2000 (38%) samples of OCT and X-Ray data respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge