Yannick Schwartz
PARIETAL, NEUROSPIN
Assessing and tuning brain decoders: cross-validation, caveats, and guidelines
Nov 07, 2016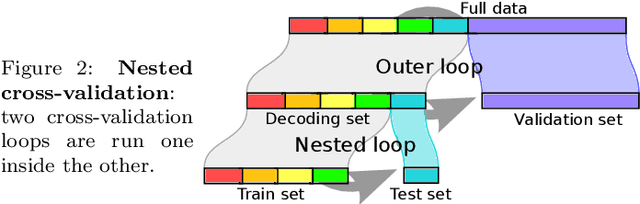
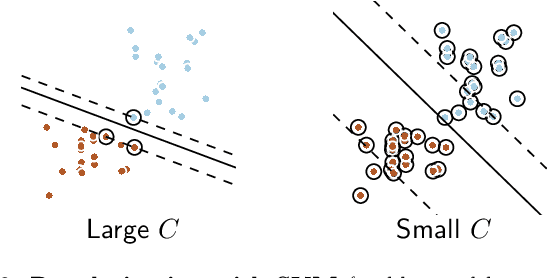
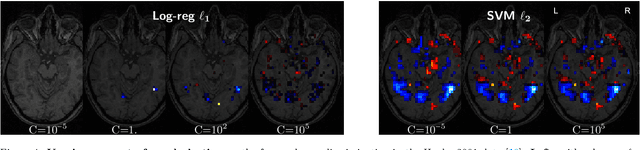

Abstract:Decoding, ie prediction from brain images or signals, calls for empirical evaluation of its predictive power. Such evaluation is achieved via cross-validation, a method also used to tune decoders' hyper-parameters. This paper is a review on cross-validation procedures for decoding in neuroimaging. It includes a didactic overview of the relevant theoretical considerations. Practical aspects are highlighted with an extensive empirical study of the common decoders in within-and across-subject predictions, on multiple datasets --anatomical and functional MRI and MEG-- and simulations. Theory and experiments outline that the popular " leave-one-out " strategy leads to unstable and biased estimates, and a repeated random splits method should be preferred. Experiments outline the large error bars of cross-validation in neuroimaging settings: typical confidence intervals of 10%. Nested cross-validation can tune decoders' parameters while avoiding circularity bias. However we find that it can be more favorable to use sane defaults, in particular for non-sparse decoders.
Mapping cognitive ontologies to and from the brain
Nov 20, 2013



Abstract:Imaging neuroscience links brain activation maps to behavior and cognition via correlational studies. Due to the nature of the individual experiments, based on eliciting neural response from a small number of stimuli, this link is incomplete, and unidirectional from the causal point of view. To come to conclusions on the function implied by the activation of brain regions, it is necessary to combine a wide exploration of the various brain functions and some inversion of the statistical inference. Here we introduce a methodology for accumulating knowledge towards a bidirectional link between observed brain activity and the corresponding function. We rely on a large corpus of imaging studies and a predictive engine. Technically, the challenges are to find commonality between the studies without denaturing the richness of the corpus. The key elements that we contribute are labeling the tasks performed with a cognitive ontology, and modeling the long tail of rare paradigms in the corpus. To our knowledge, our approach is the first demonstration of predicting the cognitive content of completely new brain images. To that end, we propose a method that predicts the experimental paradigms across different studies.
PyXNAT: XNAT in Python
Jan 29, 2013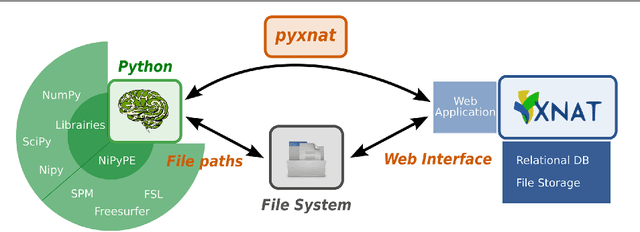


Abstract:As neuroimaging databases grow in size and complexity, the time researchers spend investigating and managing the data increases to the expense of data analysis. As a result, investigators rely more and more heavily on scripting using high-level languages to automate data management and processing tasks. For this, a structured and programmatic access to the data store is necessary. Web services are a first step toward this goal. They however lack in functionality and ease of use because they provide only low level interfaces to databases. We introduce here PyXNAT, a Python module that interacts with The Extensible Neuroimaging Archive Toolkit (XNAT) through native Python calls across multiple operating systems. The choice of Python enables PyXNAT to expose the XNAT Web Services and unify their features with a higher level and more expressive language. PyXNAT provides XNAT users direct access to all the scientific packages in Python. Finally PyXNAT aims to be efficient and easy to use, both as a backend library to build XNAT clients and as an alternative frontend from the command line.
Improving accuracy and power with transfer learning using a meta-analytic database
Sep 28, 2012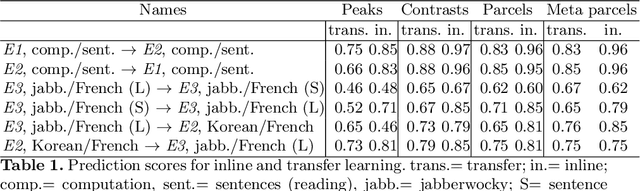
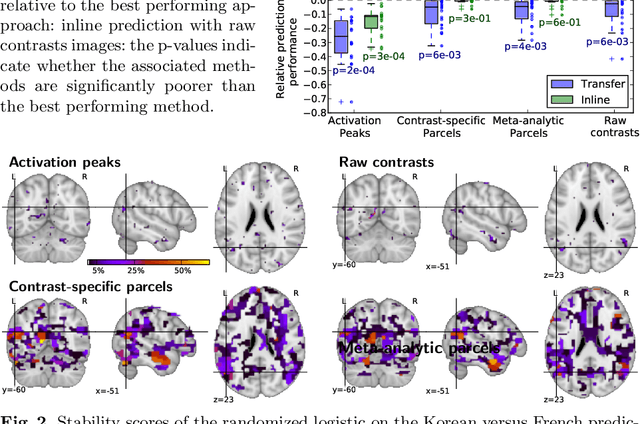
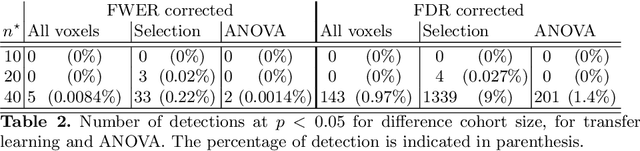
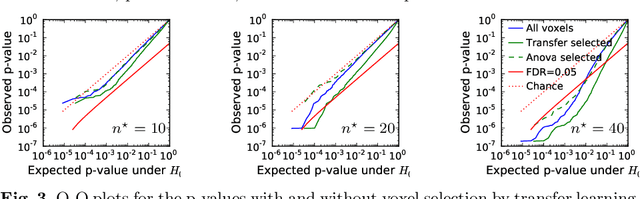
Abstract:Typical cohorts in brain imaging studies are not large enough for systematic testing of all the information contained in the images. To build testable working hypotheses, investigators thus rely on analysis of previous work, sometimes formalized in a so-called meta-analysis. In brain imaging, this approach underlies the specification of regions of interest (ROIs) that are usually selected on the basis of the coordinates of previously detected effects. In this paper, we propose to use a database of images, rather than coordinates, and frame the problem as transfer learning: learning a discriminant model on a reference task to apply it to a different but related new task. To facilitate statistical analysis of small cohorts, we use a sparse discriminant model that selects predictive voxels on the reference task and thus provides a principled procedure to define ROIs. The benefits of our approach are twofold. First it uses the reference database for prediction, i.e. to provide potential biomarkers in a clinical setting. Second it increases statistical power on the new task. We demonstrate on a set of 18 pairs of functional MRI experimental conditions that our approach gives good prediction. In addition, on a specific transfer situation involving different scanners at different locations, we show that voxel selection based on transfer learning leads to higher detection power on small cohorts.
On spatial selectivity and prediction across conditions with fMRI
Sep 07, 2012


Abstract:Researchers in functional neuroimaging mostly use activation coordinates to formulate their hypotheses. Instead, we propose to use the full statistical images to define regions of interest (ROIs). This paper presents two machine learning approaches, transfer learning and selection transfer, that are compared upon their ability to identify the common patterns between brain activation maps related to two functional tasks. We provide some preliminary quantification of these similarities, and show that selection transfer makes it possible to set a spatial scale yielding ROIs that are more specific to the context of interest than with transfer learning. In particular, selection transfer outlines well known regions such as the Visual Word Form Area when discriminating between different visual tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge