Yang Tai
Same contribution
NTIRE 2025 challenge on Text to Image Generation Model Quality Assessment
May 22, 2025Abstract:This paper reports on the NTIRE 2025 challenge on Text to Image (T2I) generation model quality assessment, which will be held in conjunction with the New Trends in Image Restoration and Enhancement Workshop (NTIRE) at CVPR 2025. The aim of this challenge is to address the fine-grained quality assessment of text-to-image generation models. This challenge evaluates text-to-image models from two aspects: image-text alignment and image structural distortion detection, and is divided into the alignment track and the structural track. The alignment track uses the EvalMuse-40K, which contains around 40K AI-Generated Images (AIGIs) generated by 20 popular generative models. The alignment track has a total of 371 registered participants. A total of 1,883 submissions are received in the development phase, and 507 submissions are received in the test phase. Finally, 12 participating teams submitted their models and fact sheets. The structure track uses the EvalMuse-Structure, which contains 10,000 AI-Generated Images (AIGIs) with corresponding structural distortion mask. A total of 211 participants have registered in the structure track. A total of 1155 submissions are received in the development phase, and 487 submissions are received in the test phase. Finally, 8 participating teams submitted their models and fact sheets. Almost all methods have achieved better results than baseline methods, and the winning methods in both tracks have demonstrated superior prediction performance on T2I model quality assessment.
EvalMuse-40K: A Reliable and Fine-Grained Benchmark with Comprehensive Human Annotations for Text-to-Image Generation Model Evaluation
Dec 24, 2024Abstract:Recently, Text-to-Image (T2I) generation models have achieved significant advancements. Correspondingly, many automated metrics have emerged to evaluate the image-text alignment capabilities of generative models. However, the performance comparison among these automated metrics is limited by existing small datasets. Additionally, these datasets lack the capacity to assess the performance of automated metrics at a fine-grained level. In this study, we contribute an EvalMuse-40K benchmark, gathering 40K image-text pairs with fine-grained human annotations for image-text alignment-related tasks. In the construction process, we employ various strategies such as balanced prompt sampling and data re-annotation to ensure the diversity and reliability of our benchmark. This allows us to comprehensively evaluate the effectiveness of image-text alignment metrics for T2I models. Meanwhile, we introduce two new methods to evaluate the image-text alignment capabilities of T2I models: FGA-BLIP2 which involves end-to-end fine-tuning of a vision-language model to produce fine-grained image-text alignment scores and PN-VQA which adopts a novel positive-negative VQA manner in VQA models for zero-shot fine-grained evaluation. Both methods achieve impressive performance in image-text alignment evaluations. We also use our methods to rank current AIGC models, in which the results can serve as a reference source for future study and promote the development of T2I generation. The data and code will be made publicly available.
A Deep Learning Based Workflow for Detection of Lung Nodules With Chest Radiograph
Dec 21, 2021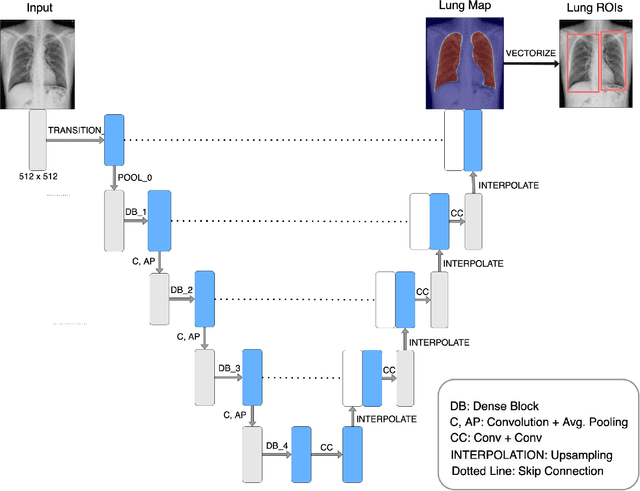
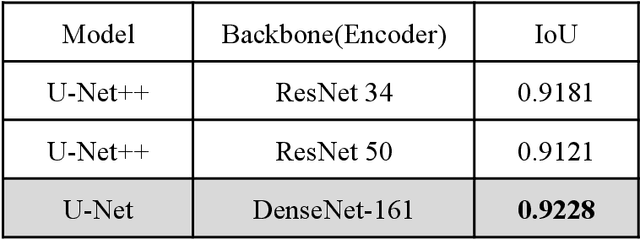
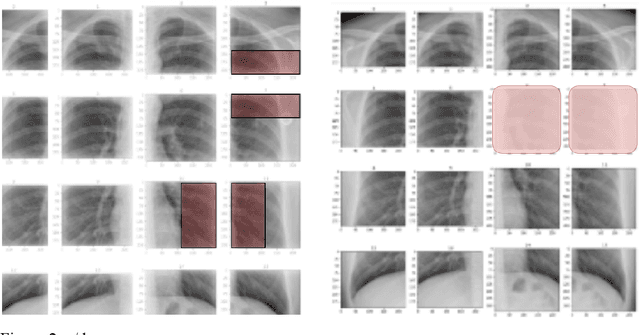
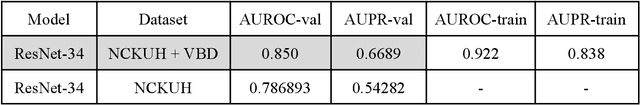
Abstract:PURPOSE: This study aimed to develop a deep learning-based tool to detect and localize lung nodules with chest radiographs(CXRs). We expected it to enhance the efficiency of interpreting CXRs and reduce the possibilities of delayed diagnosis of lung cancer. MATERIALS AND METHODS: We collected CXRs from NCKUH database and VBD, an open-source medical image dataset, as our training and validation data. A number of CXRs from the Ministry of Health and Welfare(MOHW) database served as our test data. We built a segmentation model to identify lung areas from CXRs, and sliced them into 16 patches. Physicians labeled the CXRs by clicking the patches. These labeled patches were then used to train and fine-tune a deep neural network(DNN) model, classifying the patches as positive or negative. Finally, we test the DNN model with the lung patches of CXRs from MOHW. RESULTS: Our segmentation model identified the lung regions well from the whole CXR. The Intersection over Union(IoU) between the ground truth and the segmentation result was 0.9228. In addition, our DNN model achieved a sensitivity of 0.81, specificity of 0.82, and AUROC of 0.869 in 98 of 125 cases. For the other 27 difficult cases, the sensitivity was 0.54, specificity 0.494, and AUROC 0.682. Overall, we obtained a sensitivity of 0.78, specificity of 0.79, and AUROC 0.837. CONCLUSIONS: Our two-step workflow is comparable to state-of-the-art algorithms in the sensitivity and specificity of localizing lung nodules from CXRs. Notably, our workflow provides an efficient way for specialists to label the data, which is valuable for relevant researches because of the relative rarity of labeled medical image data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge