Yair Hanani
Tensor-Parallelism with Partially Synchronized Activations
Jun 24, 2025Abstract:Training and inference of Large Language Models (LLMs) with tensor-parallelism requires substantial communication to synchronize activations. Our findings suggest that with a few minor adjustments to current practices, LLMs can be trained without fully synchronizing activations, reducing bandwidth demands. We name this "Communication-Aware Architecture for Tensor-parallelism" (CAAT-Net). We train 1B and 7B parameter CAAT-Net models, with a 50% reduction in tensor-parallel communication and no significant drop in pretraining accuracy. Furthermore, we demonstrate how CAAT-Net accelerates both training and inference workloads.
Dual Precision Quantization for Efficient and Accurate Deep Neural Networks Inference
May 20, 2025Abstract:Deep neural networks have achieved state-of-the-art results in a wide range of applications, from natural language processing and computer vision to speech recognition. However, as tasks become increasingly complex, model sizes continue to grow, posing challenges in latency and memory efficiency. To meet these constraints, post-training quantization has emerged as a promising solution. In this paper, we propose a novel hardware-efficient quantization and inference scheme that exploits hardware advantages with minimal accuracy degradation. Specifically, we introduce a W4A8 scheme, where weights are quantized and stored using 4-bit integer precision, and inference computations are performed using 8-bit floating-point arithmetic, demonstrating significant speedups and improved memory utilization compared to 16-bit operations, applicable on various modern accelerators. To mitigate accuracy loss, we develop a novel quantization algorithm, dubbed Dual Precision Quantization (DPQ), that leverages the unique structure of our scheme without introducing additional inference overhead. Experimental results demonstrate improved performance (i.e., increased throughput) while maintaining tolerable accuracy degradation relative to the full-precision model.
Automatic mixed precision for optimizing gained time with constrained loss mean-squared-error based on model partition to sequential sub-graphs
May 19, 2025Abstract:Quantization is essential for Neural Network (NN) compression, reducing model size and computational demands by using lower bit-width data types, though aggressive reduction often hampers accuracy. Mixed Precision (MP) mitigates this tradeoff by varying the numerical precision across network layers. This study focuses on automatically selecting an optimal MP configuration within Post-Training Quantization (PTQ) for inference. The first key contribution is a novel sensitivity metric derived from a first-order Taylor series expansion of the loss function as a function of quantization errors in weights and activations. This metric, based on the Mean Square Error (MSE) of the loss, is efficiently calculated per layer using high-precision forward and backward passes over a small calibration dataset. The metric is additive across layers, with low calibration memory overhead as weight optimization is unnecessary. The second contribution is an accurate hardware-aware method for predicting MP time gain by modeling it as additive for sequential sub-graphs. An algorithm partitions the model graph into sequential subgraphs, measuring time gain for each configuration using a few samples. After calibrating per-layer sensitivity and time gain, an Integer Programming (IP) problem is formulated to maximize time gain while keeping loss MSE below a set threshold. Memory gain and theoretical time gain based on Multiply and Accumulate (MAC) operations are also considered. Rigorous experiments on the Intel Gaudi 2 accelerator validate the approach on several Large Language Models (LLMs).
Improving Post Training Neural Quantization: Layer-wise Calibration and Integer Programming
Jun 14, 2020



Abstract:Most of the literature on neural network quantization requires some training of the quantized model (fine-tuning). However, this training is not always possible in real-world scenarios, as it requires the full dataset. Lately, post-training quantization methods have gained considerable attention, as they are simple to use and require only a small, unlabeled calibration set. Yet, they usually incur significant accuracy degradation when quantized below 8-bits. This paper seeks to address this problem by introducing two pipelines, advanced and light, where the former involves: (i) minimizing the quantization errors of each layer by optimizing its parameters over the calibration set; (ii) using integer programming to optimally allocate the desired bit-width for each layer while constraining accuracy degradation or model compression; and (iii) tuning the mixed-precision model statistics to correct biases introduced during quantization. While the light pipeline which invokes only (ii) and (iii) obtains surprisingly accurate results; the advanced pipeline yields state-of-the-art accuracy-compression ratios for both vision and text models. For instance, on ResNet50, we obtain less than 1% accuracy degradation while compressing the model to 13% of its original size. We open-sourced our code.
DeepGestalt - Identifying Rare Genetic Syndromes Using Deep Learning
Jan 23, 2018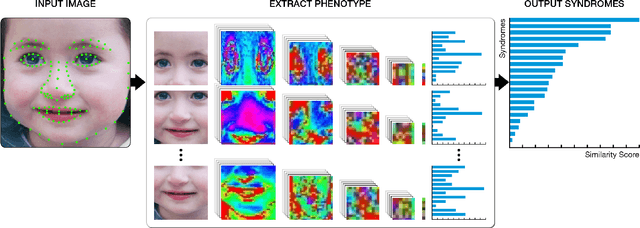
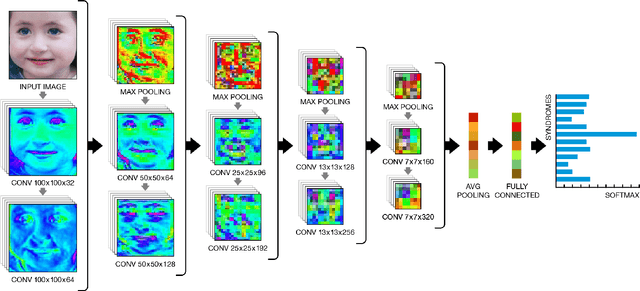

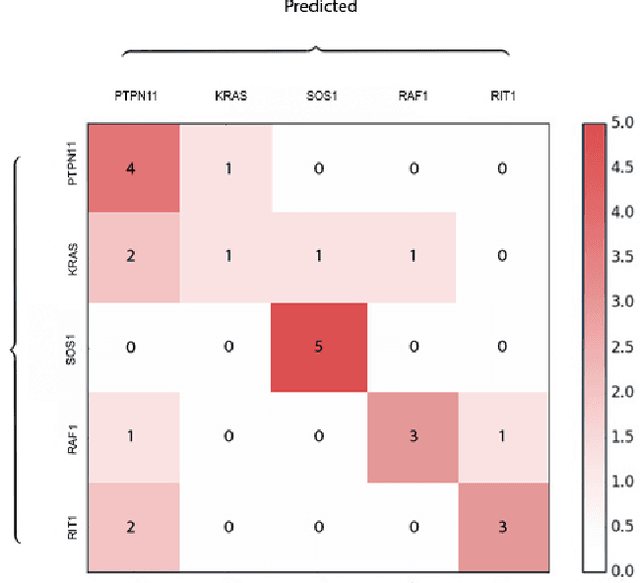
Abstract:Facial analysis technologies have recently measured up to the capabilities of expert clinicians in syndrome identification. To date, these technologies could only identify phenotypes of a few diseases, limiting their role in clinical settings where hundreds of diagnoses must be considered. We developed a facial analysis framework, DeepGestalt, using computer vision and deep learning algorithms, that quantifies similarities to hundreds of genetic syndromes based on unconstrained 2D images. DeepGestalt is currently trained with over 26,000 patient cases from a rapidly growing phenotype-genotype database, consisting of tens of thousands of validated clinical cases, curated through a community-driven platform. DeepGestalt currently achieves 91% top-10-accuracy in identifying over 215 different genetic syndromes and has outperformed clinical experts in three separate experiments. We suggest that this form of artificial intelligence is ready to support medical genetics in clinical and laboratory practices and will play a key role in the future of precision medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge