Yaron Gurovich
On Disentangled and Locally Fair Representations
May 05, 2022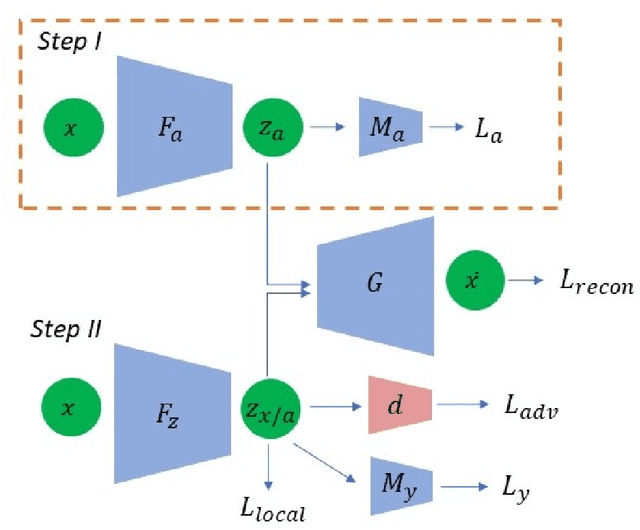
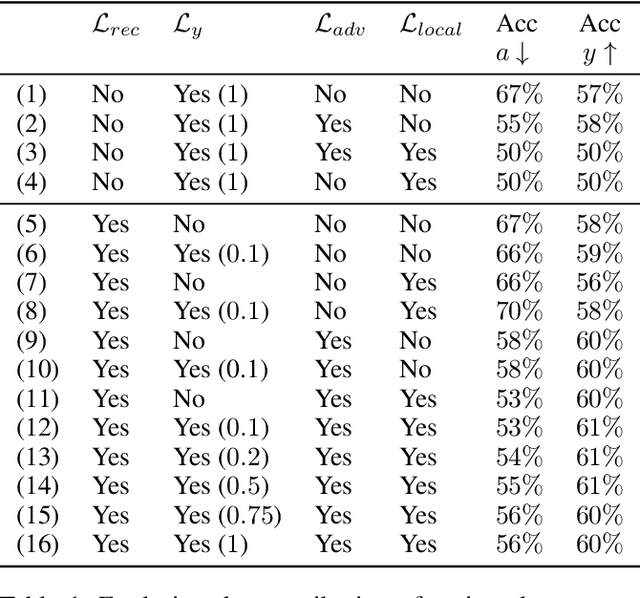
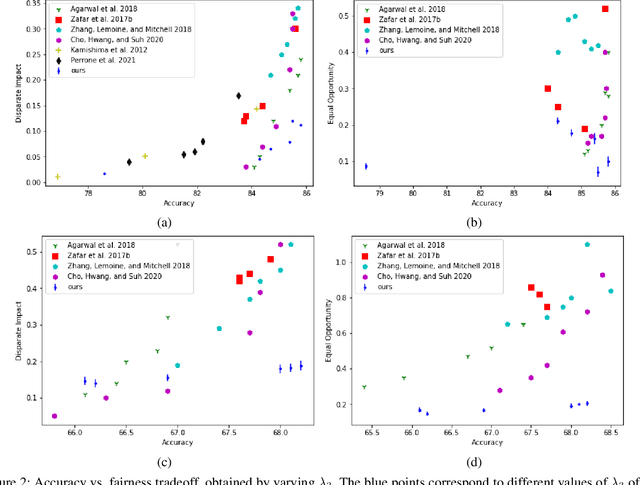
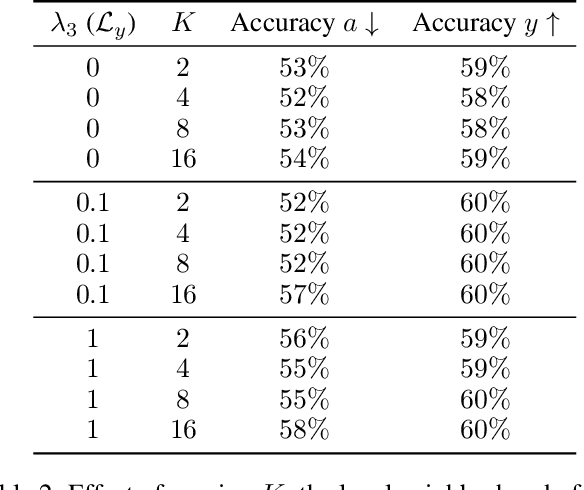
Abstract:We study the problem of performing classification in a manner that is fair for sensitive groups, such as race and gender. This problem is tackled through the lens of disentangled and locally fair representations. We learn a locally fair representation, such that, under the learned representation, the neighborhood of each sample is balanced in terms of the sensitive attribute. For instance, when a decision is made to hire an individual, we ensure that the $K$ most similar hired individuals are racially balanced. Crucially, we ensure that similar individuals are found based on attributes not correlated to their race. To this end, we disentangle the embedding space into two representations. The first of which is correlated with the sensitive attribute while the second is not. We apply our local fairness objective only to the second, uncorrelated, representation. Through a set of experiments, we demonstrate the necessity of both disentangled and local fairness for obtaining fair and accurate representations. We evaluate our method on real-world settings such as predicting income and re-incarceration rate and demonstrate the advantage of our method.
DeepGestalt - Identifying Rare Genetic Syndromes Using Deep Learning
Jan 23, 2018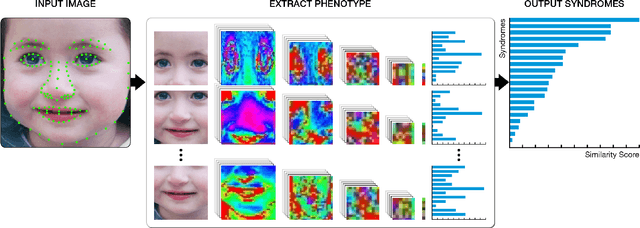
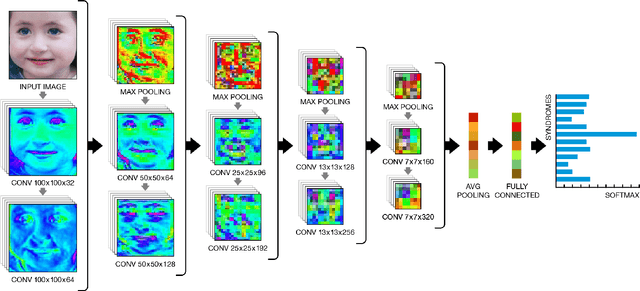

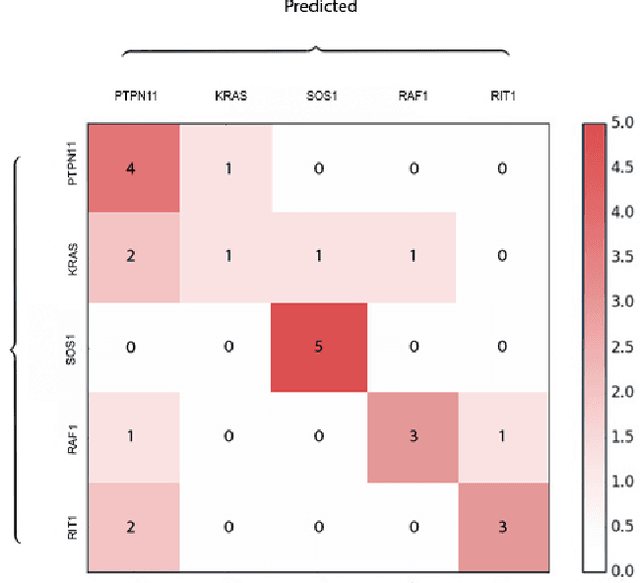
Abstract:Facial analysis technologies have recently measured up to the capabilities of expert clinicians in syndrome identification. To date, these technologies could only identify phenotypes of a few diseases, limiting their role in clinical settings where hundreds of diagnoses must be considered. We developed a facial analysis framework, DeepGestalt, using computer vision and deep learning algorithms, that quantifies similarities to hundreds of genetic syndromes based on unconstrained 2D images. DeepGestalt is currently trained with over 26,000 patient cases from a rapidly growing phenotype-genotype database, consisting of tens of thousands of validated clinical cases, curated through a community-driven platform. DeepGestalt currently achieves 91% top-10-accuracy in identifying over 215 different genetic syndromes and has outperformed clinical experts in three separate experiments. We suggest that this form of artificial intelligence is ready to support medical genetics in clinical and laboratory practices and will play a key role in the future of precision medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge