Xuexue Sun
Moore Threads
Pre-trained Encoder Inference: Revealing Upstream Encoders In Downstream Machine Learning Services
Aug 05, 2024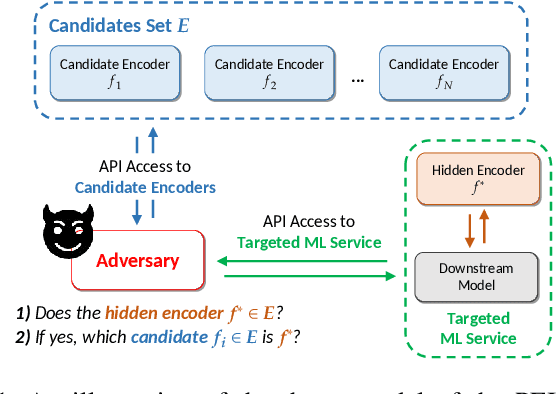

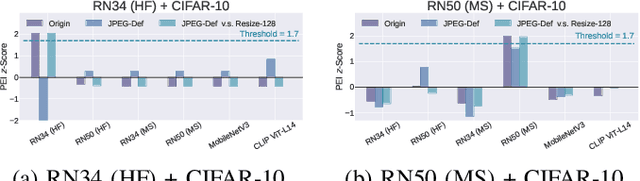
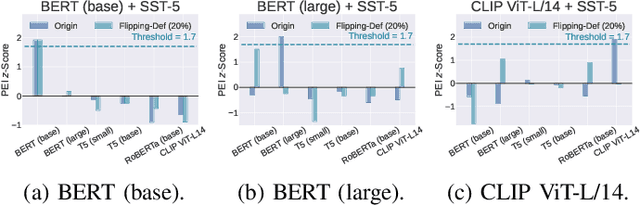
Abstract:Though pre-trained encoders can be easily accessed online to build downstream machine learning (ML) services quickly, various attacks have been designed to compromise the security and privacy of these encoders. While most attacks target encoders on the upstream side, it remains unknown how an encoder could be threatened when deployed in a downstream ML service. This paper unveils a new vulnerability: the Pre-trained Encoder Inference (PEI) attack, which posts privacy threats toward encoders hidden behind downstream ML services. By only providing API accesses to a targeted downstream service and a set of candidate encoders, the PEI attack can infer which encoder is secretly used by the targeted service based on candidate ones. We evaluate the attack performance of PEI against real-world encoders on three downstream tasks: image classification, text classification, and text-to-image generation. Experiments show that the PEI attack succeeds in revealing the hidden encoder in most cases and seldom makes mistakes even when the hidden encoder is not in the candidate set. We also conducted a case study on one of the most recent vision-language models, LLaVA, to illustrate that the PEI attack is useful in assisting other ML attacks such as adversarial attacks. The code is available at https://github.com/fshp971/encoder-inference.
Advanced Multi-Microscopic Views Cell Semi-supervised Segmentation
Mar 21, 2023Abstract:Although deep learning (DL) shows powerful potential in cell segmentation tasks, it suffers from poor generalization as DL-based methods originally simplified cell segmentation in detecting cell membrane boundary, lacking prominent cellular structures to position overall differentiating. Moreover, the scarcity of annotated cell images limits the performance of DL models. Segmentation limitations of a single category of cell make massive practice difficult, much less, with varied modalities. In this paper, we introduce a novel semi-supervised cell segmentation method called Multi-Microscopic-view Cell semi-supervised Segmentation (MMCS), which can train cell segmentation models utilizing less labeled multi-posture cell images with different microscopy well. Technically, MMCS consists of Nucleus-assisted global recognition, Self-adaptive diameter filter, and Temporal-ensembling models. Nucleus-assisted global recognition adds additional cell nucleus channel to improve the global distinguishing performance of fuzzy cell membrane boundaries even when cells aggregate. Besides, self-adapted cell diameter filter can help separate multi-resolution cells with different morphology properly. It further leverages the temporal-ensembling models to improve the semi-supervised training process, achieving effective training with less labeled data. Additionally, optimizing the weight of unlabeled loss contributed to total loss also improve the model performance. Evaluated on the Tuning Set of NeurIPS 2022 Cell Segmentation Challenge (NeurIPS CellSeg), MMCS achieves an F1-score of 0.8239 and the running time for all cases is within the time tolerance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge