Xuanting Xie
Attention Beyond Neighborhoods: Reviving Transformer for Graph Clustering
Sep 18, 2025Abstract:Attention mechanisms have become a cornerstone in modern neural networks, driving breakthroughs across diverse domains. However, their application to graph structured data, where capturing topological connections is essential, remains underexplored and underperforming compared to Graph Neural Networks (GNNs), particularly in the graph clustering task. GNN tends to overemphasize neighborhood aggregation, leading to a homogenization of node representations. Conversely, Transformer tends to over globalize, highlighting distant nodes at the expense of meaningful local patterns. This dichotomy raises a key question: Is attention inherently redundant for unsupervised graph learning? To address this, we conduct a comprehensive empirical analysis, uncovering the complementary weaknesses of GNN and Transformer in graph clustering. Motivated by these insights, we propose the Attentive Graph Clustering Network (AGCN) a novel architecture that reinterprets the notion that graph is attention. AGCN directly embeds the attention mechanism into the graph structure, enabling effective global information extraction while maintaining sensitivity to local topological cues. Our framework incorporates theoretical analysis to contrast AGCN behavior with GNN and Transformer and introduces two innovations: (1) a KV cache mechanism to improve computational efficiency, and (2) a pairwise margin contrastive loss to boost the discriminative capacity of the attention space. Extensive experimental results demonstrate that AGCN outperforms state-of-the-art methods.
Aggregation-aware MLP: An Unsupervised Approach for Graph Message-passing
Jul 27, 2025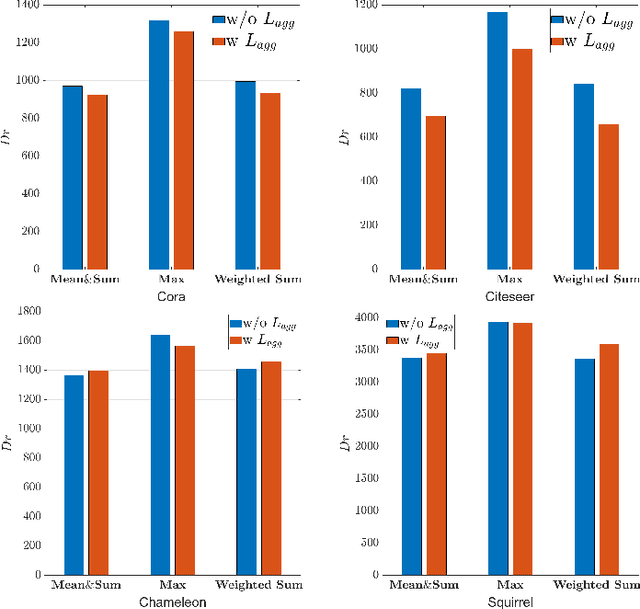
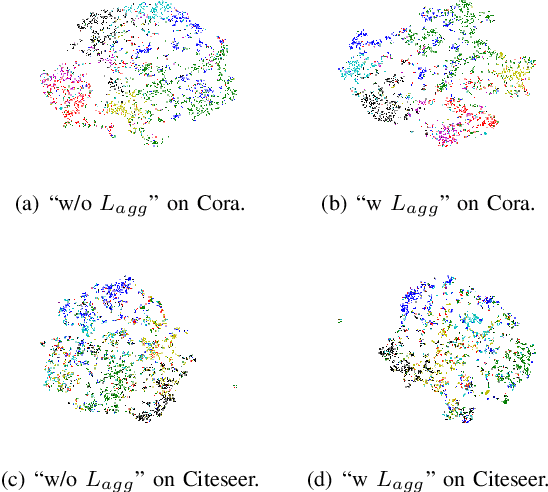
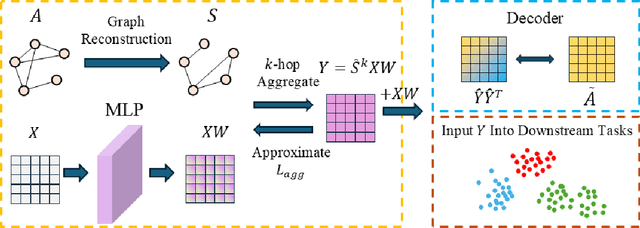
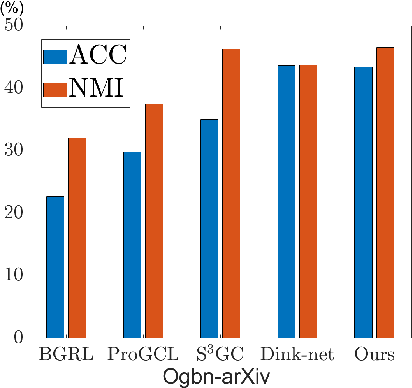
Abstract:Graph Neural Networks (GNNs) have become a dominant approach to learning graph representations, primarily because of their message-passing mechanisms. However, GNNs typically adopt a fixed aggregator function such as Mean, Max, or Sum without principled reasoning behind the selection. This rigidity, especially in the presence of heterophily, often leads to poor, problem dependent performance. Although some attempts address this by designing more sophisticated aggregation functions, these methods tend to rely heavily on labeled data, which is often scarce in real-world tasks. In this work, we propose a novel unsupervised framework, "Aggregation-aware Multilayer Perceptron" (AMLP), which shifts the paradigm from directly crafting aggregation functions to making MLP adaptive to aggregation. Our lightweight approach consists of two key steps: First, we utilize a graph reconstruction method that facilitates high-order grouping effects, and second, we employ a single-layer network to encode varying degrees of heterophily, thereby improving the capacity and applicability of the model. Extensive experiments on node clustering and classification demonstrate the superior performance of AMLP, highlighting its potential for diverse graph learning scenarios.
One Node One Model: Featuring the Missing-Half for Graph Clustering
Dec 13, 2024



Abstract:Most existing graph clustering methods primarily focus on exploiting topological structure, often neglecting the ``missing-half" node feature information, especially how these features can enhance clustering performance. This issue is further compounded by the challenges associated with high-dimensional features. Feature selection in graph clustering is particularly difficult because it requires simultaneously discovering clusters and identifying the relevant features for these clusters. To address this gap, we introduce a novel paradigm called ``one node one model", which builds an exclusive model for each node and defines the node label as a combination of predictions for node groups. Specifically, the proposed ``Feature Personalized Graph Clustering (FPGC)" method identifies cluster-relevant features for each node using a squeeze-and-excitation block, integrating these features into each model to form the final representations. Additionally, the concept of feature cross is developed as a data augmentation technique to learn low-order feature interactions. Extensive experimental results demonstrate that FPGC outperforms state-of-the-art clustering methods. Moreover, the plug-and-play nature of our method provides a versatile solution to enhance GNN-based models from a feature perspective.
Provable Filter for Real-world Graph Clustering
Mar 06, 2024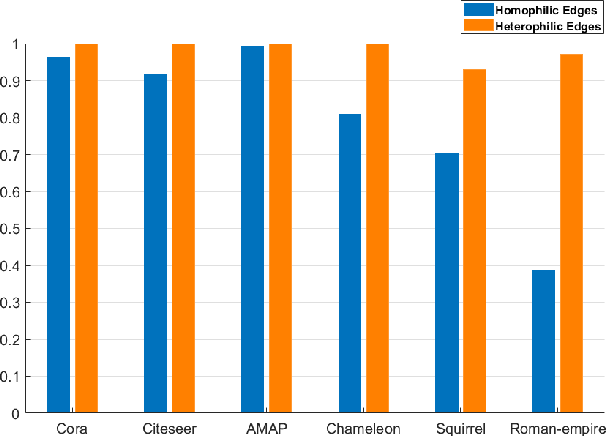
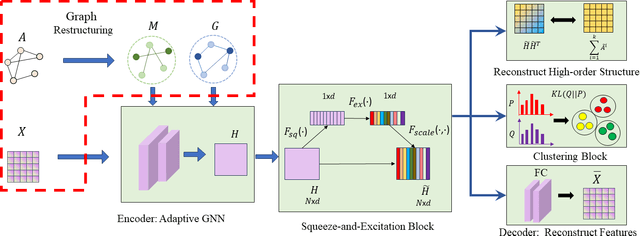
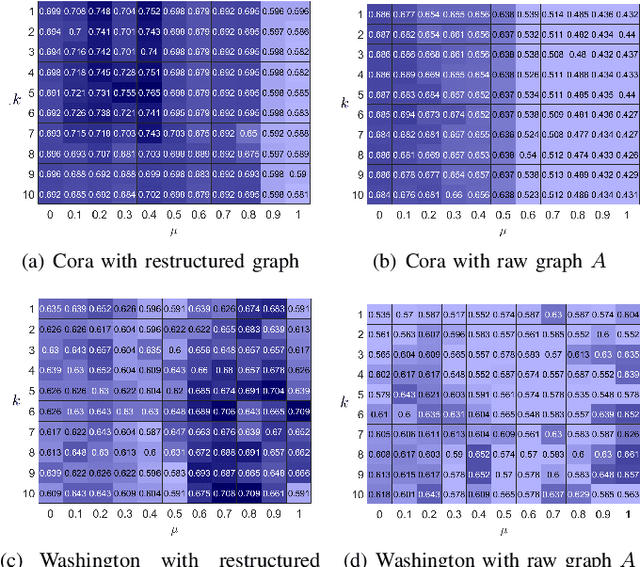
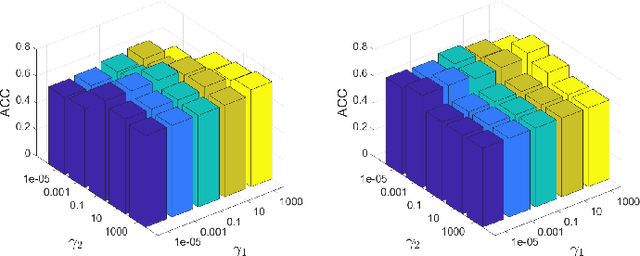
Abstract:Graph clustering, an important unsupervised problem, has been shown to be more resistant to advances in Graph Neural Networks (GNNs). In addition, almost all clustering methods focus on homophilic graphs and ignore heterophily. This significantly limits their applicability in practice, since real-world graphs exhibit a structural disparity and cannot simply be classified as homophily and heterophily. Thus, a principled way to handle practical graphs is urgently needed. To fill this gap, we provide a novel solution with theoretical support. Interestingly, we find that most homophilic and heterophilic edges can be correctly identified on the basis of neighbor information. Motivated by this finding, we construct two graphs that are highly homophilic and heterophilic, respectively. They are used to build low-pass and high-pass filters to capture holistic information. Important features are further enhanced by the squeeze-and-excitation block. We validate our approach through extensive experiments on both homophilic and heterophilic graphs. Empirical results demonstrate the superiority of our method compared to state-of-the-art clustering methods.
Simplified PCNet with Robustness
Mar 06, 2024
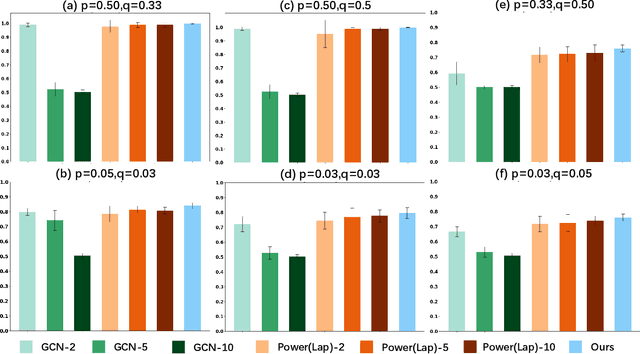
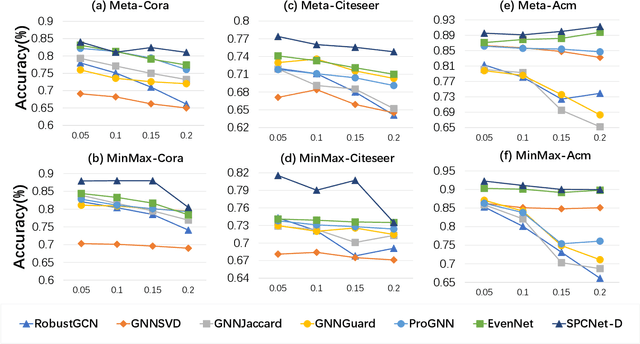
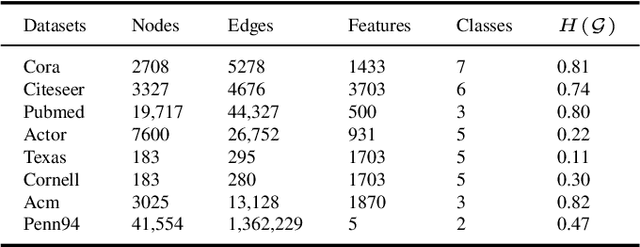
Abstract:Graph Neural Networks (GNNs) have garnered significant attention for their success in learning the representation of homophilic or heterophilic graphs. However, they cannot generalize well to real-world graphs with different levels of homophily. In response, the Possion-Charlier Network (PCNet) \cite{li2024pc}, the previous work, allows graph representation to be learned from heterophily to homophily. Although PCNet alleviates the heterophily issue, there remain some challenges in further improving the efficacy and efficiency. In this paper, we simplify PCNet and enhance its robustness. We first extend the filter order to continuous values and reduce its parameters. Two variants with adaptive neighborhood sizes are implemented. Theoretical analysis shows our model's robustness to graph structure perturbations or adversarial attacks. We validate our approach through semi-supervised learning tasks on various datasets representing both homophilic and heterophilic graphs.
CDC: A Simple Framework for Complex Data Clustering
Mar 06, 2024



Abstract:In today's data-driven digital era, the amount as well as complexity, such as multi-view, non-Euclidean, and multi-relational, of the collected data are growing exponentially or even faster. Clustering, which unsupervisely extracts valid knowledge from data, is extremely useful in practice. However, existing methods are independently developed to handle one particular challenge at the expense of the others. In this work, we propose a simple but effective framework for complex data clustering (CDC) that can efficiently process different types of data with linear complexity. We first utilize graph filtering to fuse geometry structure and attribute information. We then reduce the complexity with high-quality anchors that are adaptively learned via a novel similarity-preserving regularizer. We illustrate the cluster-ability of our proposed method theoretically and experimentally. In particular, we deploy CDC to graph data of size 111M.
Robust Graph Structure Learning under Heterophily
Mar 06, 2024Abstract:Graph is a fundamental mathematical structure in characterizing relations between different objects and has been widely used on various learning tasks. Most methods implicitly assume a given graph to be accurate and complete. However, real data is inevitably noisy and sparse, which will lead to inferior results. Despite the remarkable success of recent graph representation learning methods, they inherently presume that the graph is homophilic, and largely overlook heterophily, where most connected nodes are from different classes. In this regard, we propose a novel robust graph structure learning method to achieve a high-quality graph from heterophilic data for downstream tasks. We first apply a high-pass filter to make each node more distinctive from its neighbors by encoding structure information into the node features. Then, we learn a robust graph with an adaptive norm characterizing different levels of noise. Afterwards, we propose a novel regularizer to further refine the graph structure. Clustering and semi-supervised classification experiments on heterophilic graphs verify the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge