Xingtong Wang
RetCompletion:High-Speed Inference Image Completion with Retentive Network
Oct 05, 2024



Abstract:Time cost is a major challenge in achieving high-quality pluralistic image completion. Recently, the Retentive Network (RetNet) in natural language processing offers a novel approach to this problem with its low-cost inference capabilities. Inspired by this, we apply RetNet to the pluralistic image completion task in computer vision. We present RetCompletion, a two-stage framework. In the first stage, we introduce Bi-RetNet, a bidirectional sequence information fusion model that integrates contextual information from images. During inference, we employ a unidirectional pixel-wise update strategy to restore consistent image structures, achieving both high reconstruction quality and fast inference speed. In the second stage, we use a CNN for low-resolution upsampling to enhance texture details. Experiments on ImageNet and CelebA-HQ demonstrate that our inference speed is 10$\times$ faster than ICT and 15$\times$ faster than RePaint. The proposed RetCompletion significantly improves inference speed and delivers strong performance, especially when masks cover large areas of the image.
Segmentation Network with Compound Loss Function for Hydatidiform Mole Hydrops Lesion Recognition
Apr 11, 2022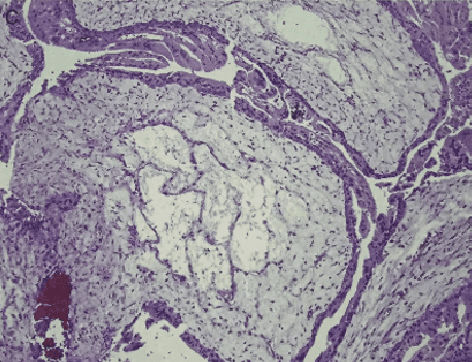
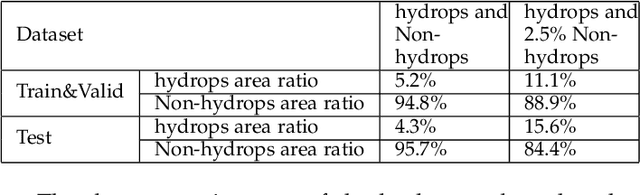
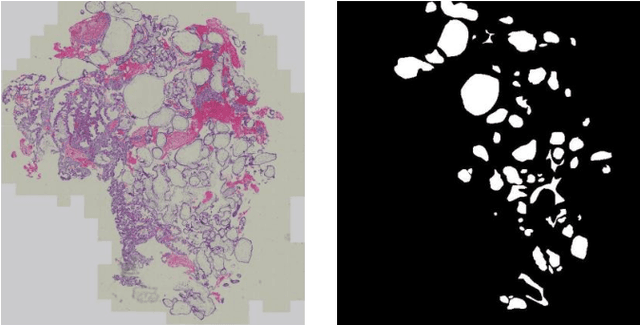
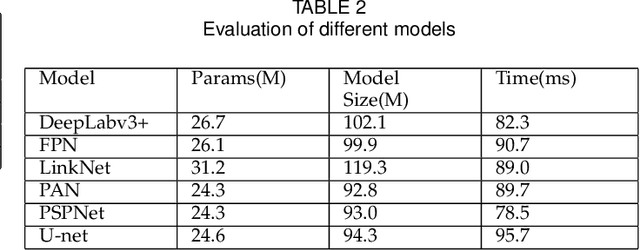
Abstract:Pathological morphology diagnosis is the standard diagnosis method of hydatidiform mole. As a disease with malignant potential, the hydatidiform mole section of hydrops lesions is an important basis for diagnosis. Due to incomplete lesion development, early hydatidiform mole is difficult to distinguish, resulting in a low accuracy of clinical diagnosis. As a remarkable machine learning technology, image semantic segmentation networks have been used in many medical image recognition tasks. We developed a hydatidiform mole hydrops lesion segmentation model based on a novel loss function and training method. The model consists of different networks that segment the section image at the pixel and lesion levels. Our compound loss function assign weights to the segmentation results of the two levels to calculate the loss. We then propose a stagewise training method to combine the advantages of various loss functions at different levels. We evaluate our method on a hydatidiform mole hydrops dataset. Experiments show that the proposed model with our loss function and training method has good recognition performance under different segmentation metrics.
A Semantic Segmentation Network Based Real-Time Computer-Aided Diagnosis System for Hydatidiform Mole Hydrops Lesion Recognition in Microscopic View
Apr 11, 2022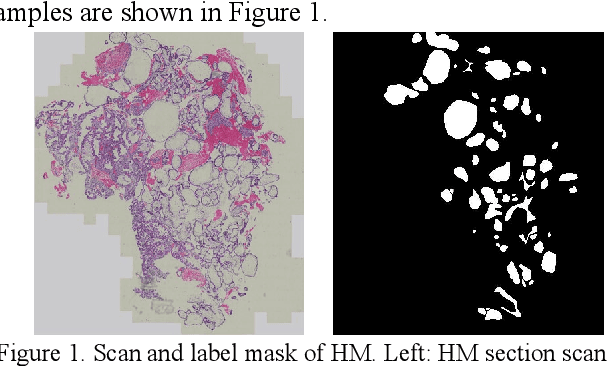
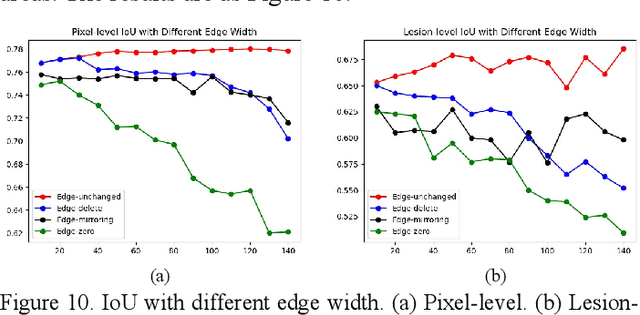
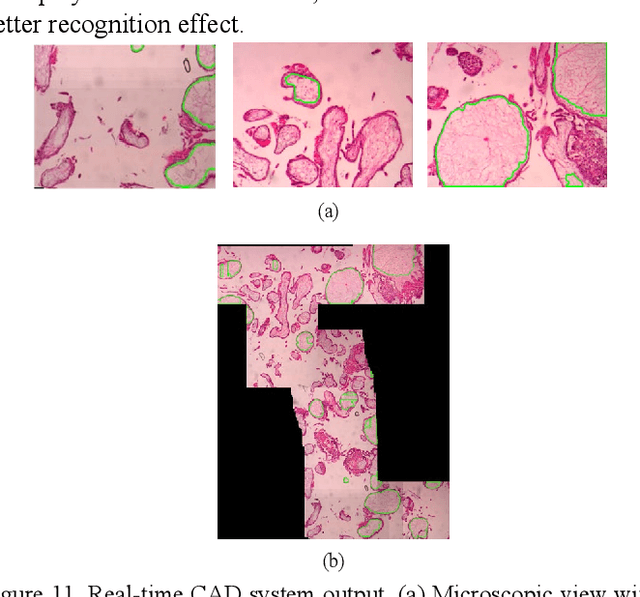
Abstract:As a disease with malignant potential, hydatidiform mole (HM) is one of the most common gestational trophoblastic diseases. For pathologists, the HM section of hydrops lesions is an important basis for diagnosis. In pathology departments, the diverse microscopic manifestations of HM lesions and the limited view under the microscope mean that physicians with extensive diagnostic experience are required to prevent missed diagnosis and misdiagnosis. Feature extraction can significantly improve the accuracy and speed of the diagnostic process. As a remarkable diagnosis assisting technology, computer-aided diagnosis (CAD) has been widely used in clinical practice. We constructed a deep-learning-based CAD system to identify HM hydrops lesions in the microscopic view in real-time. The system consists of three modules; the image mosaic module and edge extension module process the image to improve the outcome of the hydrops lesion recognition module, which adopts a semantic segmentation network, our novel compound loss function, and a stepwise training function in order to achieve the best performance in identifying hydrops lesions. We evaluated our system using an HM hydrops dataset. Experiments show that our system is able to respond in real-time and correctly display the entire microscopic view with accurately labeled HM hydrops lesions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge