Zehua Ji
SMAP: A Novel Heterogeneous Information Framework for Scenario-based Optimal Model Assignment
May 23, 2023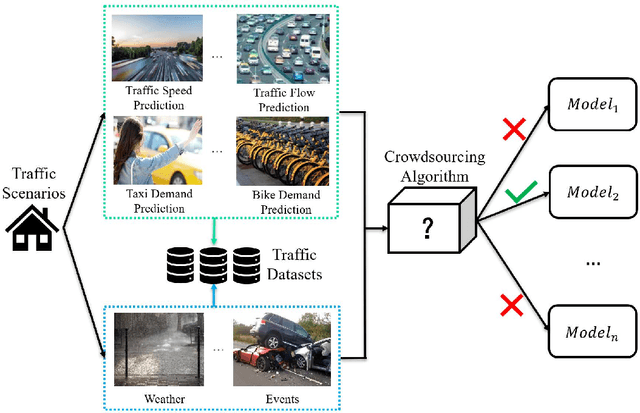
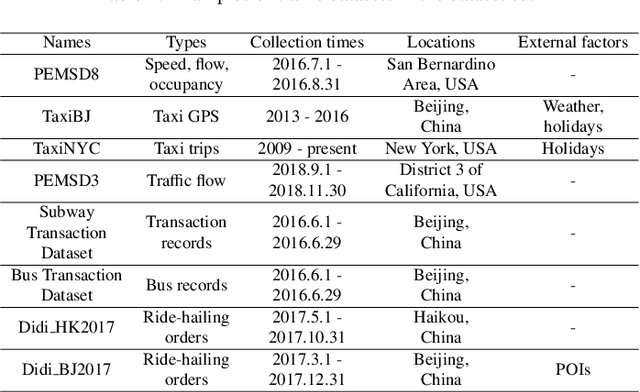
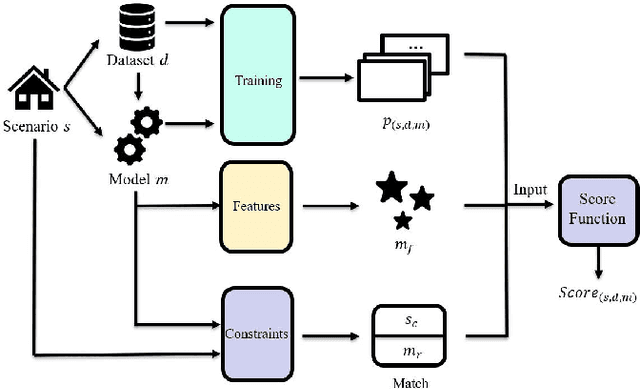
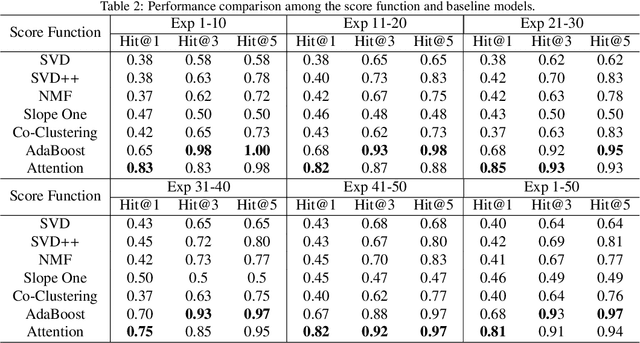
Abstract:The increasing maturity of big data applications has led to a proliferation of models targeting the same objectives within the same scenarios and datasets. However, selecting the most suitable model that considers model's features while taking specific requirements and constraints into account still poses a significant challenge. Existing methods have focused on worker-task assignments based on crowdsourcing, they neglect the scenario-dataset-model assignment problem. To address this challenge, a new problem named the Scenario-based Optimal Model Assignment (SOMA) problem is introduced and a novel framework entitled Scenario and Model Associative percepts (SMAP) is developed. SMAP is a heterogeneous information framework that can integrate various types of information to intelligently select a suitable dataset and allocate the optimal model for a specific scenario. To comprehensively evaluate models, a new score function that utilizes multi-head attention mechanisms is proposed. Moreover, a novel memory mechanism named the mnemonic center is developed to store the matched heterogeneous information and prevent duplicate matching. Six popular traffic scenarios are selected as study cases and extensive experiments are conducted on a dataset to verify the effectiveness and efficiency of SMAP and the score function.
Segmentation Network with Compound Loss Function for Hydatidiform Mole Hydrops Lesion Recognition
Apr 11, 2022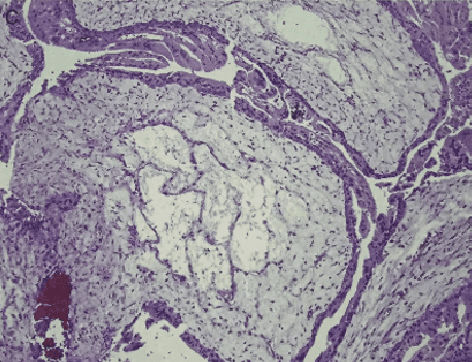
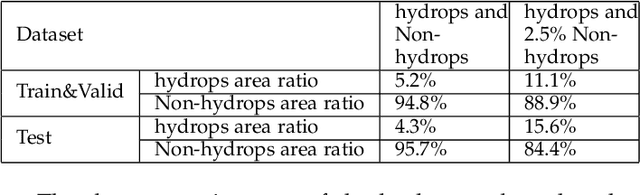
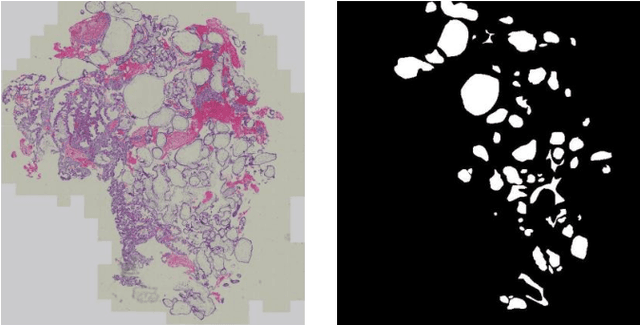
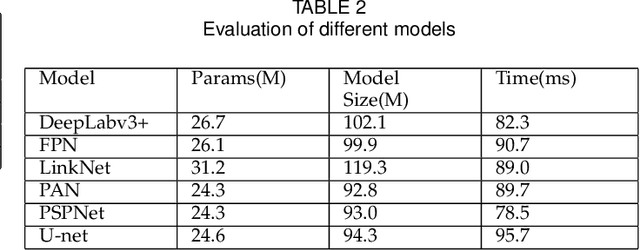
Abstract:Pathological morphology diagnosis is the standard diagnosis method of hydatidiform mole. As a disease with malignant potential, the hydatidiform mole section of hydrops lesions is an important basis for diagnosis. Due to incomplete lesion development, early hydatidiform mole is difficult to distinguish, resulting in a low accuracy of clinical diagnosis. As a remarkable machine learning technology, image semantic segmentation networks have been used in many medical image recognition tasks. We developed a hydatidiform mole hydrops lesion segmentation model based on a novel loss function and training method. The model consists of different networks that segment the section image at the pixel and lesion levels. Our compound loss function assign weights to the segmentation results of the two levels to calculate the loss. We then propose a stagewise training method to combine the advantages of various loss functions at different levels. We evaluate our method on a hydatidiform mole hydrops dataset. Experiments show that the proposed model with our loss function and training method has good recognition performance under different segmentation metrics.
A Semantic Segmentation Network Based Real-Time Computer-Aided Diagnosis System for Hydatidiform Mole Hydrops Lesion Recognition in Microscopic View
Apr 11, 2022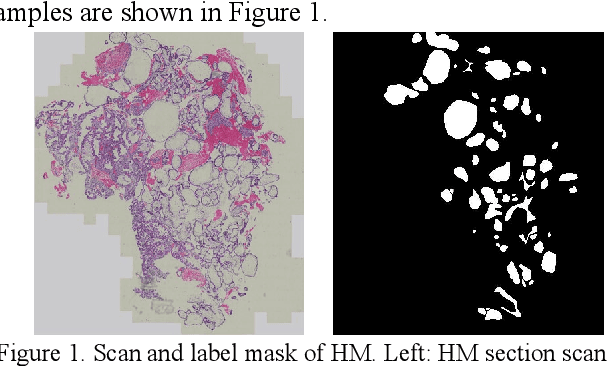
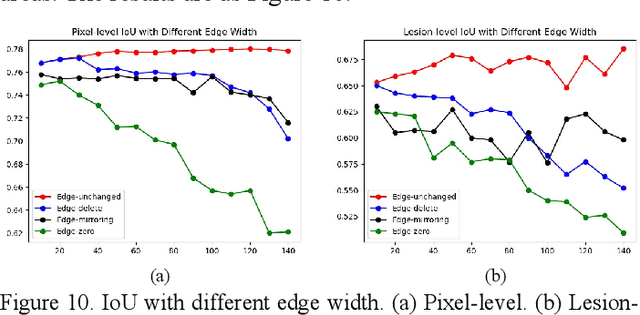
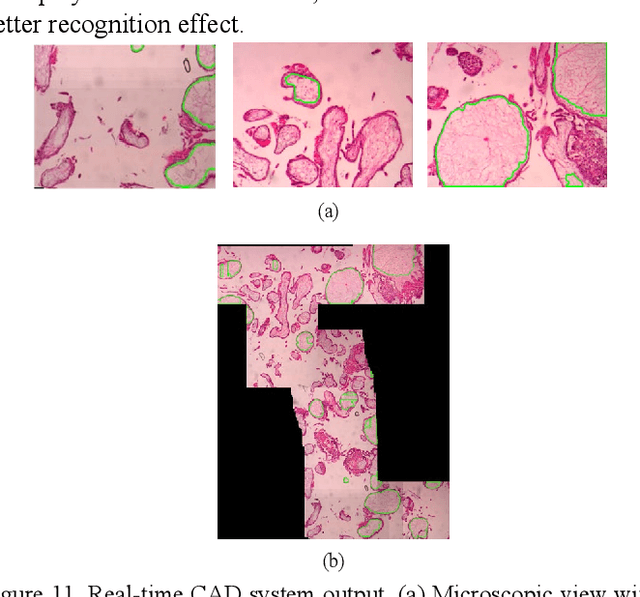
Abstract:As a disease with malignant potential, hydatidiform mole (HM) is one of the most common gestational trophoblastic diseases. For pathologists, the HM section of hydrops lesions is an important basis for diagnosis. In pathology departments, the diverse microscopic manifestations of HM lesions and the limited view under the microscope mean that physicians with extensive diagnostic experience are required to prevent missed diagnosis and misdiagnosis. Feature extraction can significantly improve the accuracy and speed of the diagnostic process. As a remarkable diagnosis assisting technology, computer-aided diagnosis (CAD) has been widely used in clinical practice. We constructed a deep-learning-based CAD system to identify HM hydrops lesions in the microscopic view in real-time. The system consists of three modules; the image mosaic module and edge extension module process the image to improve the outcome of the hydrops lesion recognition module, which adopts a semantic segmentation network, our novel compound loss function, and a stepwise training function in order to achieve the best performance in identifying hydrops lesions. We evaluated our system using an HM hydrops dataset. Experiments show that our system is able to respond in real-time and correctly display the entire microscopic view with accurately labeled HM hydrops lesions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge