Xingran Xie
LoFormer: Local Frequency Transformer for Image Deblurring
Jul 24, 2024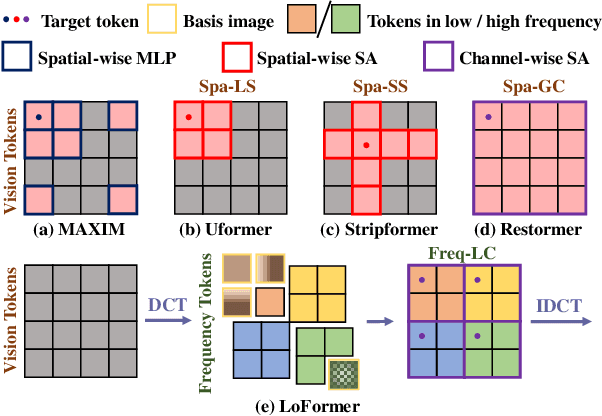
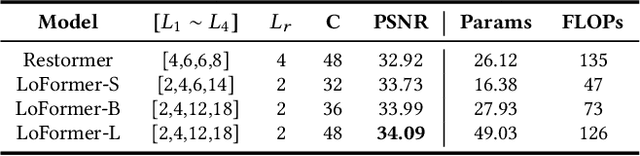
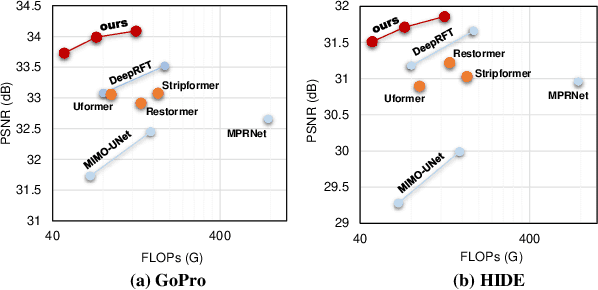
Abstract:Due to the computational complexity of self-attention (SA), prevalent techniques for image deblurring often resort to either adopting localized SA or employing coarse-grained global SA methods, both of which exhibit drawbacks such as compromising global modeling or lacking fine-grained correlation. In order to address this issue by effectively modeling long-range dependencies without sacrificing fine-grained details, we introduce a novel approach termed Local Frequency Transformer (LoFormer). Within each unit of LoFormer, we incorporate a Local Channel-wise SA in the frequency domain (Freq-LC) to simultaneously capture cross-covariance within low- and high-frequency local windows. These operations offer the advantage of (1) ensuring equitable learning opportunities for both coarse-grained structures and fine-grained details, and (2) exploring a broader range of representational properties compared to coarse-grained global SA methods. Additionally, we introduce an MLP Gating mechanism complementary to Freq-LC, which serves to filter out irrelevant features while enhancing global learning capabilities. Our experiments demonstrate that LoFormer significantly improves performance in the image deblurring task, achieving a PSNR of 34.09 dB on the GoPro dataset with 126G FLOPs. https://github.com/DeepMed-Lab-ECNU/Single-Image-Deblur
Gene-induced Multimodal Pre-training for Image-omic Classification
Sep 06, 2023Abstract:Histology analysis of the tumor micro-environment integrated with genomic assays is the gold standard for most cancers in modern medicine. This paper proposes a Gene-induced Multimodal Pre-training (GiMP) framework, which jointly incorporates genomics and Whole Slide Images (WSIs) for classification tasks. Our work aims at dealing with the main challenges of multi-modality image-omic classification w.r.t. (1) the patient-level feature extraction difficulties from gigapixel WSIs and tens of thousands of genes, and (2) effective fusion considering high-order relevance modeling. Concretely, we first propose a group multi-head self-attention gene encoder to capture global structured features in gene expression cohorts. We design a masked patch modeling paradigm (MPM) to capture the latent pathological characteristics of different tissues. The mask strategy is randomly masking a fixed-length contiguous subsequence of patch embeddings of a WSI. Finally, we combine the classification tokens of paired modalities and propose a triplet learning module to learn high-order relevance and discriminative patient-level information.After pre-training, a simple fine-tuning can be adopted to obtain the classification results. Experimental results on the TCGA dataset show the superiority of our network architectures and our pre-training framework, achieving 99.47% in accuracy for image-omic classification. The code is publicly available at https://github.com/huangwudiduan/GIMP.
S$^3$R: Self-supervised Spectral Regression for Hyperspectral Histopathology Image Classification
Sep 19, 2022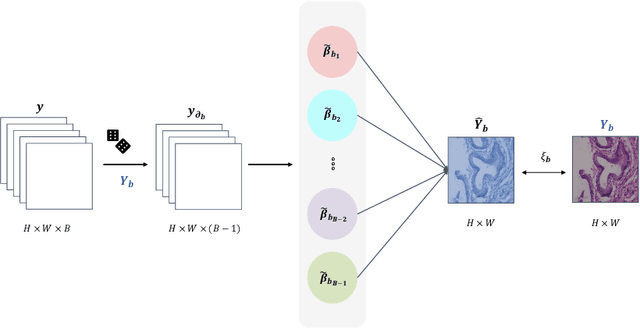
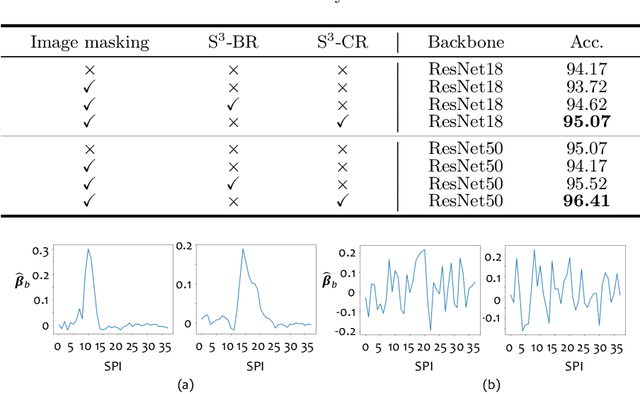
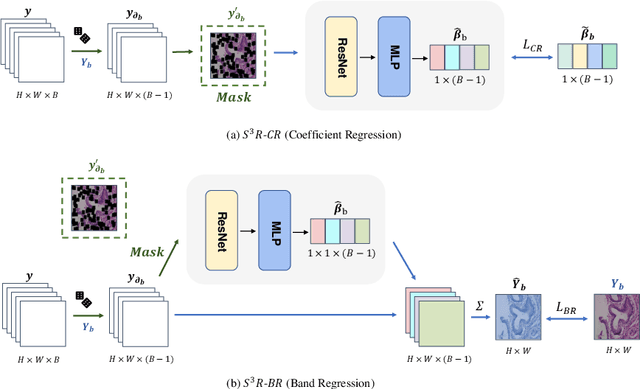
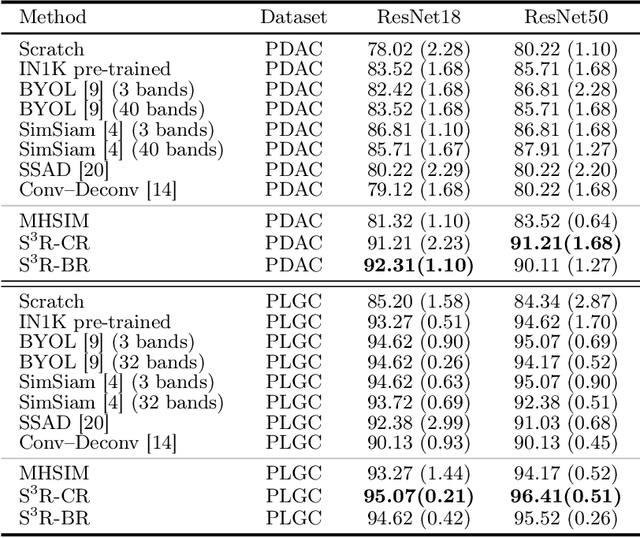
Abstract:Benefited from the rich and detailed spectral information in hyperspectral images (HSI), HSI offers great potential for a wide variety of medical applications such as computational pathology. But, the lack of adequate annotated data and the high spatiospectral dimensions of HSIs usually make classification networks prone to overfit. Thus, learning a general representation which can be transferred to the downstream tasks is imperative. To our knowledge, no appropriate self-supervised pre-training method has been designed for histopathology HSIs. In this paper, we introduce an efficient and effective Self-supervised Spectral Regression (S$^3$R) method, which exploits the low rank characteristic in the spectral domain of HSI. More concretely, we propose to learn a set of linear coefficients that can be used to represent one band by the remaining bands via masking out these bands. Then, the band is restored by using the learned coefficients to reweight the remaining bands. Two pre-text tasks are designed: (1)S$^3$R-CR, which regresses the linear coefficients, so that the pre-trained model understands the inherent structures of HSIs and the pathological characteristics of different morphologies; (2)S$^3$R-BR, which regresses the missing band, making the model to learn the holistic semantics of HSIs. Compared to prior arts i.e., contrastive learning methods, which focuses on natural images, S$^3$R converges at least 3 times faster, and achieves significant improvements up to 14% in accuracy when transferring to HSI classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge