S$^3$R: Self-supervised Spectral Regression for Hyperspectral Histopathology Image Classification
Paper and Code
Sep 19, 2022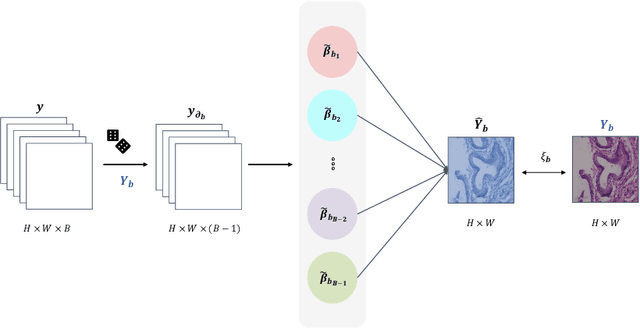
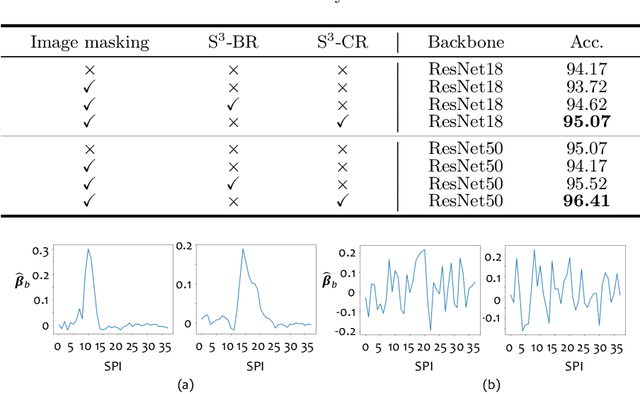
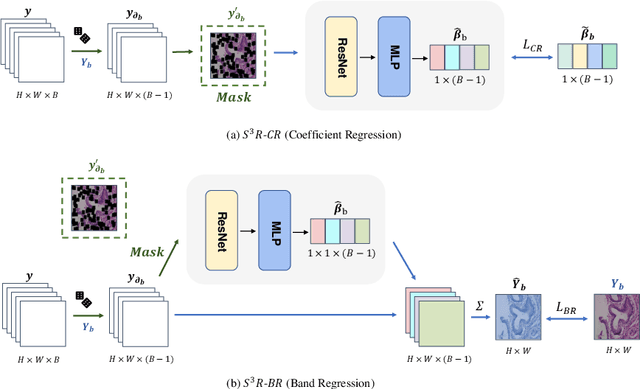
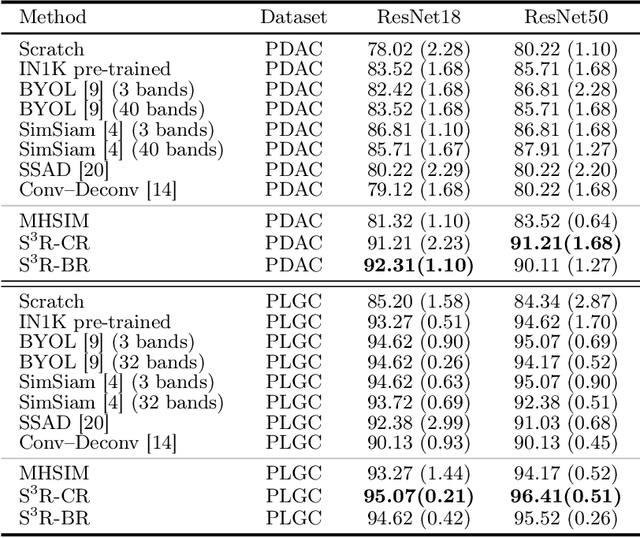
Benefited from the rich and detailed spectral information in hyperspectral images (HSI), HSI offers great potential for a wide variety of medical applications such as computational pathology. But, the lack of adequate annotated data and the high spatiospectral dimensions of HSIs usually make classification networks prone to overfit. Thus, learning a general representation which can be transferred to the downstream tasks is imperative. To our knowledge, no appropriate self-supervised pre-training method has been designed for histopathology HSIs. In this paper, we introduce an efficient and effective Self-supervised Spectral Regression (S$^3$R) method, which exploits the low rank characteristic in the spectral domain of HSI. More concretely, we propose to learn a set of linear coefficients that can be used to represent one band by the remaining bands via masking out these bands. Then, the band is restored by using the learned coefficients to reweight the remaining bands. Two pre-text tasks are designed: (1)S$^3$R-CR, which regresses the linear coefficients, so that the pre-trained model understands the inherent structures of HSIs and the pathological characteristics of different morphologies; (2)S$^3$R-BR, which regresses the missing band, making the model to learn the holistic semantics of HSIs. Compared to prior arts i.e., contrastive learning methods, which focuses on natural images, S$^3$R converges at least 3 times faster, and achieves significant improvements up to 14% in accuracy when transferring to HSI classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge