Xiaoxian Yang
MSV-Mamba: A Multiscale Vision Mamba Network for Echocardiography Segmentation
Jan 13, 2025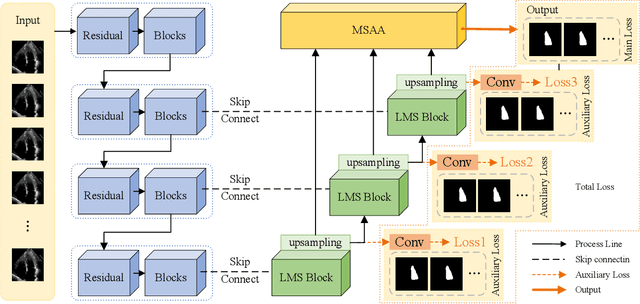
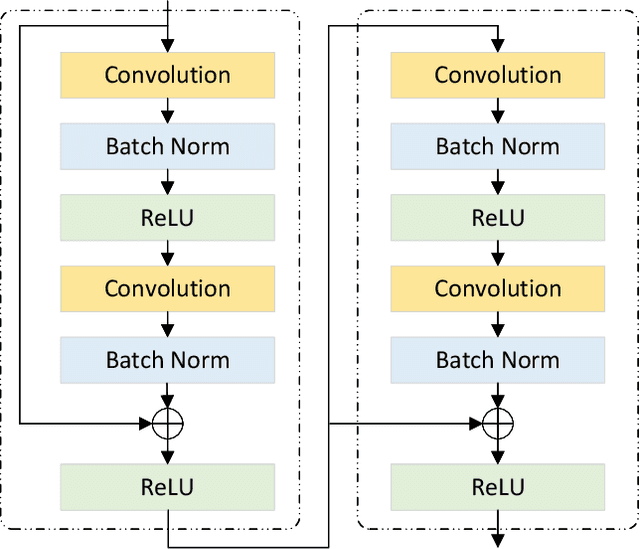
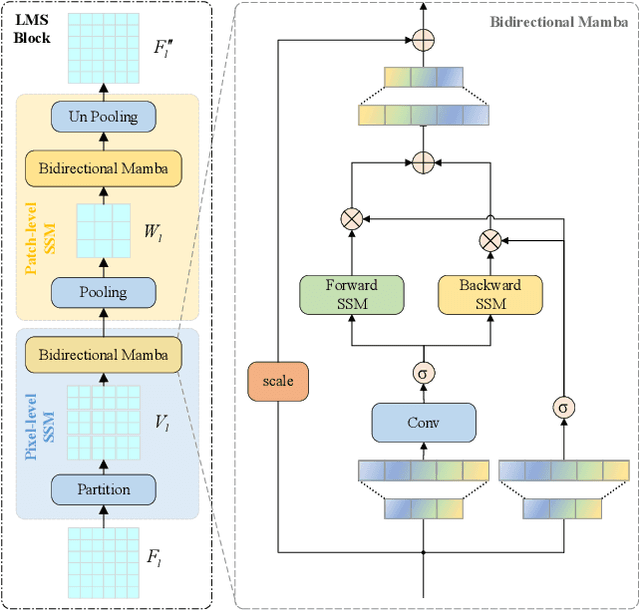
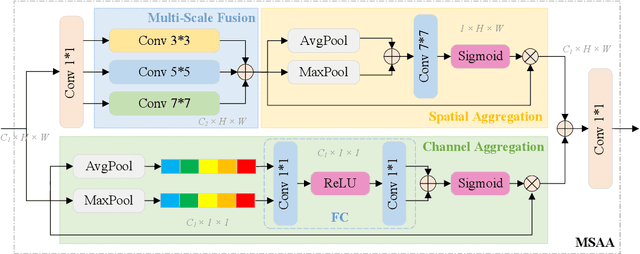
Abstract:Ultrasound imaging frequently encounters challenges, such as those related to elevated noise levels, diminished spatiotemporal resolution, and the complexity of anatomical structures. These factors significantly hinder the model's ability to accurately capture and analyze structural relationships and dynamic patterns across various regions of the heart. Mamba, an emerging model, is one of the most cutting-edge approaches that is widely applied to diverse vision and language tasks. To this end, this paper introduces a U-shaped deep learning model incorporating a large-window Mamba scale (LMS) module and a hierarchical feature fusion approach for echocardiographic segmentation. First, a cascaded residual block serves as an encoder and is employed to incrementally extract multiscale detailed features. Second, a large-window multiscale mamba module is integrated into the decoder to capture global dependencies across regions and enhance the segmentation capability for complex anatomical structures. Furthermore, our model introduces auxiliary losses at each decoder layer and employs a dual attention mechanism to fuse multilayer features both spatially and across channels. This approach enhances segmentation performance and accuracy in delineating complex anatomical structures. Finally, the experimental results using the EchoNet-Dynamic and CAMUS datasets demonstrate that the model outperforms other methods in terms of both accuracy and robustness. For the segmentation of the left ventricular endocardium (${LV}_{endo}$), the model achieved optimal values of 95.01 and 93.36, respectively, while for the left ventricular epicardium (${LV}_{epi}$), values of 87.35 and 87.80, respectively, were achieved. This represents an improvement ranging between 0.54 and 1.11 compared with the best-performing model.
Identifying Electrocardiogram Abnormalities Using a Handcrafted-Rule-Enhanced Neural Network
Jun 16, 2022



Abstract:A large number of people suffer from life-threatening cardiac abnormalities, and electrocardiogram (ECG) analysis is beneficial to determining whether an individual is at risk of such abnormalities. Automatic ECG classification methods, especially the deep learning based ones, have been proposed to detect cardiac abnormalities using ECG records, showing good potential to improve clinical diagnosis and help early prevention of cardiovascular diseases. However, the predictions of the known neural networks still do not satisfactorily meet the needs of clinicians, and this phenomenon suggests that some information used in clinical diagnosis may not be well captured and utilized by these methods. In this paper, we introduce some rules into convolutional neural networks, which help present clinical knowledge to deep learning based ECG analysis, in order to improve automated ECG diagnosis performance. Specifically, we propose a Handcrafted-Rule-enhanced Neural Network (called HRNN) for ECG classification with standard 12-lead ECG input, which consists of a rule inference module and a deep learning module. Experiments on two large-scale public ECG datasets show that our new approach considerably outperforms existing state-of-the-art methods. Further, our proposed approach not only can improve the diagnosis performance, but also can assist in detecting mislabelled ECG samples. Our codes are available at https://github.com/alwaysbyx/ecg_processing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge