Xianling Cong
Data and Knowledge Co-driving for Cancer Subtype Classification on Multi-Scale Histopathological Slides
Apr 18, 2023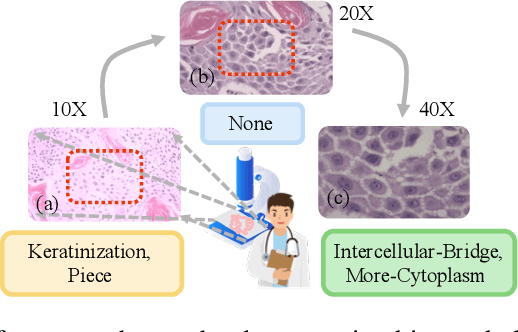
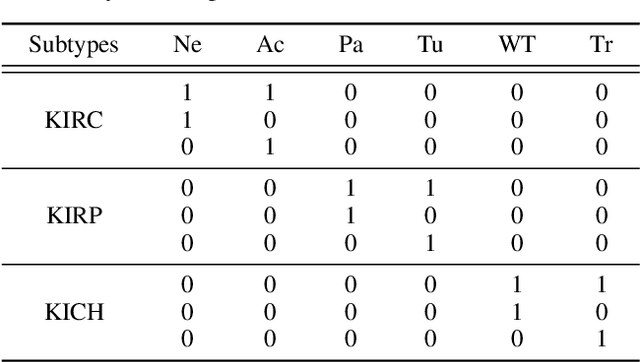
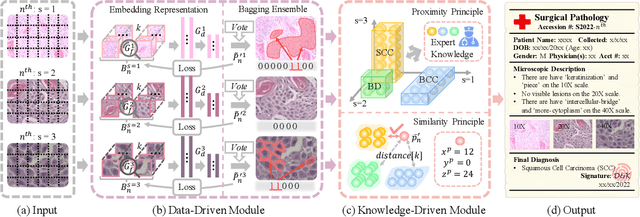
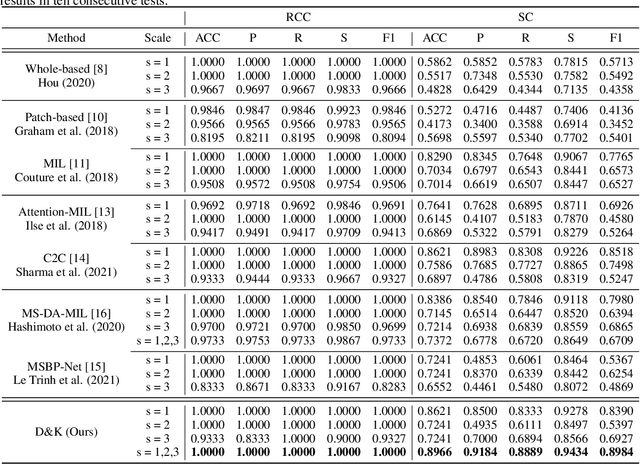
Abstract:Artificial intelligence-enabled histopathological data analysis has become a valuable assistant to the pathologist. However, existing models lack representation and inference abilities compared with those of pathologists, especially in cancer subtype diagnosis, which is unconvincing in clinical practice. For instance, pathologists typically observe the lesions of a slide from global to local, and then can give a diagnosis based on their knowledge and experience. In this paper, we propose a Data and Knowledge Co-driving (D&K) model to replicate the process of cancer subtype classification on a histopathological slide like a pathologist. Specifically, in the data-driven module, the bagging mechanism in ensemble learning is leveraged to integrate the histological features from various bags extracted by the embedding representation unit. Furthermore, a knowledge-driven module is established based on the Gestalt principle in psychology to build the three-dimensional (3D) expert knowledge space and map histological features into this space for metric. Then, the diagnosis can be made according to the Euclidean distance between them. Extensive experimental results on both public and in-house datasets demonstrate that the D&K model has a high performance and credible results compared with the state-of-the-art methods for diagnosing histopathological subtypes. Code: https://github.com/Dennis-YB/Data-and-Knowledge-Co-driving-for-Cancer-Subtypes-Classification
Multi-Modality Multi-Scale Cardiovascular Disease Subtypes Classification Using Raman Image and Medical History
Apr 18, 2023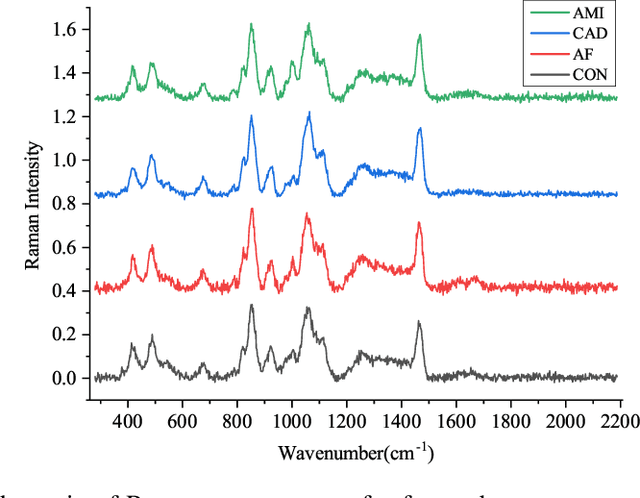
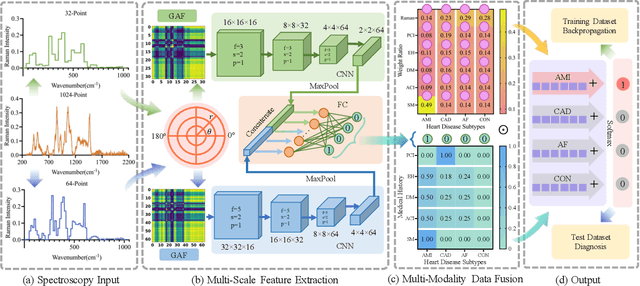
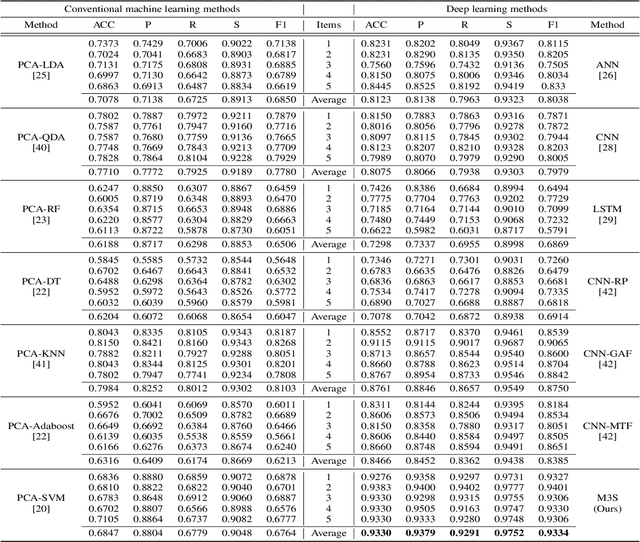
Abstract:Raman spectroscopy (RS) has been widely used for disease diagnosis, e.g., cardiovascular disease (CVD), owing to its efficiency and component-specific testing capabilities. A series of popular deep learning methods have recently been introduced to learn nuance features from RS for binary classifications and achieved outstanding performance than conventional machine learning methods. However, these existing deep learning methods still confront some challenges in classifying subtypes of CVD. For example, the nuance between subtypes is quite hard to capture and represent by intelligent models due to the chillingly similar shape of RS sequences. Moreover, medical history information is an essential resource for distinguishing subtypes, but they are underutilized. In light of this, we propose a multi-modality multi-scale model called M3S, which is a novel deep learning method with two core modules to address these issues. First, we convert RS data to various resolution images by the Gramian angular field (GAF) to enlarge nuance, and a two-branch structure is leveraged to get embeddings for distinction in the multi-scale feature extraction module. Second, a probability matrix and a weight matrix are used to enhance the classification capacity by combining the RS and medical history data in the multi-modality data fusion module. We perform extensive evaluations of M3S and found its outstanding performance on our in-house dataset, with accuracy, precision, recall, specificity, and F1 score of 0.9330, 0.9379, 0.9291, 0.9752, and 0.9334, respectively. These results demonstrate that the M3S has high performance and robustness compared with popular methods in diagnosing CVD subtypes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge