Wilson Gregory
GeometricImageNet: Extending convolutional neural networks to vector and tensor images
May 21, 2023



Abstract:Convolutional neural networks and their ilk have been very successful for many learning tasks involving images. These methods assume that the input is a scalar image representing the intensity in each pixel, possibly in multiple channels for color images. In natural-science domains however, image-like data sets might have vectors (velocity, say), tensors (polarization, say), pseudovectors (magnetic field, say), or other geometric objects in each pixel. Treating the components of these objects as independent channels in a CNN neglects their structure entirely. Our formulation -- the GeometricImageNet -- combines a geometric generalization of convolution with outer products, tensor index contractions, and tensor index permutations to construct geometric-image functions of geometric images that use and benefit from the tensor structure. The framework permits, with a very simple adjustment, restriction to function spaces that are exactly equivariant to translations, discrete rotations, and reflections. We use representation theory to quantify the dimension of the space of equivariant polynomial functions on 2-dimensional vector images. We give partial results on the expressivity of GeometricImageNet on small images. In numerical experiments, we find that GeometricImageNet has good generalization for a small simulated physics system, even when trained with a small training set. We expect this tool will be valuable for scientific and engineering machine learning, for example in cosmology or ocean dynamics.
MarkerMap: nonlinear marker selection for single-cell studies
Jul 28, 2022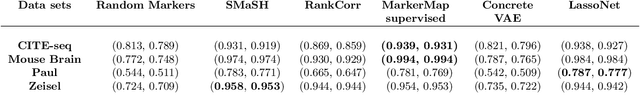
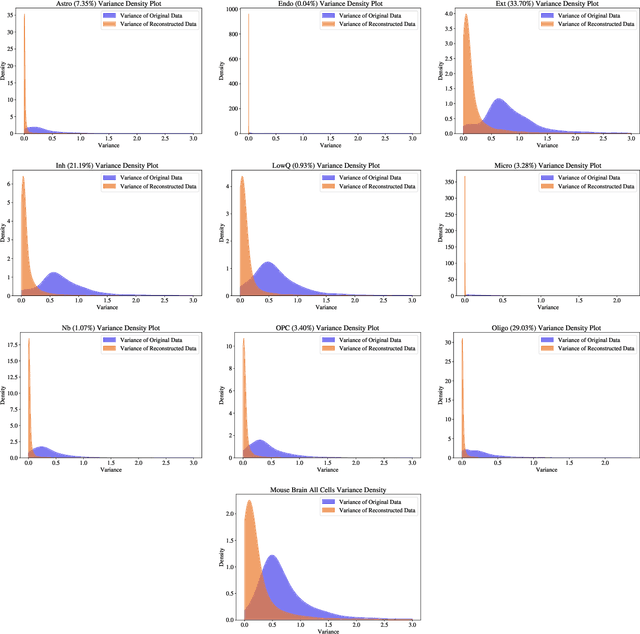
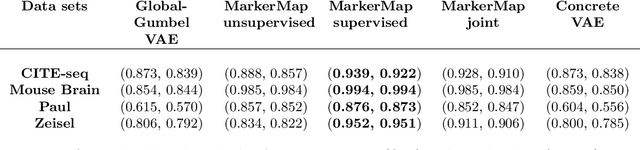
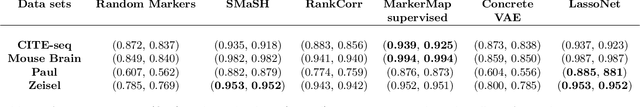
Abstract:Single-cell RNA-seq data allow the quantification of cell type differences across a growing set of biological contexts. However, pinpointing a small subset of genomic features explaining this variability can be ill-defined and computationally intractable. Here we introduce MarkerMap, a generative model for selecting minimal gene sets which are maximally informative of cell type origin and enable whole transcriptome reconstruction. MarkerMap provides a scalable framework for both supervised marker selection, aimed at identifying specific cell type populations, and unsupervised marker selection, aimed at gene expression imputation and reconstruction. We benchmark MarkerMap's competitive performance against previously published approaches on real single cell gene expression data sets. MarkerMap is available as a pip installable package, as a community resource aimed at developing explainable machine learning techniques for enhancing interpretability in single-cell studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge