Van-Khoa Le
From slides (through tiles) to pixels: an explainability framework for weakly supervised models in pre-clinical pathology
Feb 03, 2023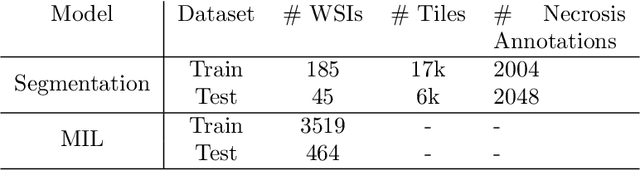
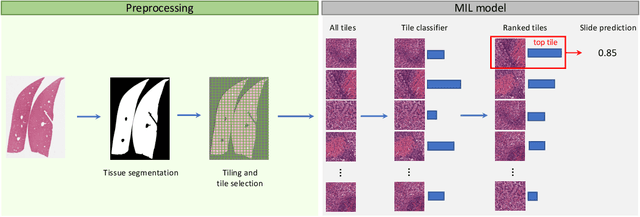
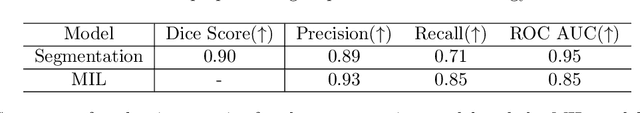
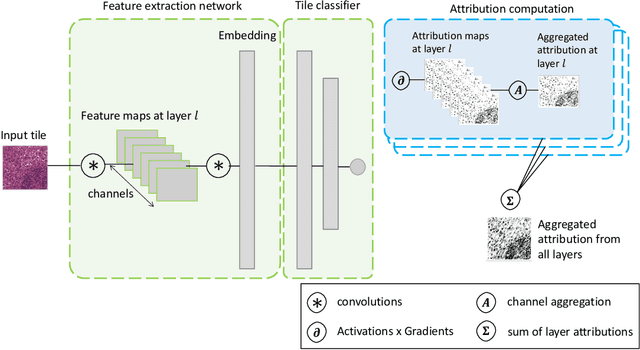
Abstract:In pre-clinical pathology, there is a paradox between the abundance of raw data (whole slide images from many organs of many individual animals) and the lack of pixel-level slide annotations done by pathologists. Due to time constraints and requirements from regulatory authorities, diagnoses are instead stored as slide labels. Weakly supervised training is designed to take advantage of those data, and the trained models can be used by pathologists to rank slides by their probability of containing a given lesion of interest. In this work, we propose a novel contextualized eXplainable AI (XAI) framework and its application to deep learning models trained on Whole Slide Images (WSIs) in Digital Pathology. Specifically, we apply our methods to a multi-instance-learning (MIL) model, which is trained solely on slide-level labels, without the need for pixel-level annotations. We validate quantitatively our methods by quantifying the agreements of our explanations' heatmaps with pathologists' annotations, as well as with predictions from a segmentation model trained on such annotations. We demonstrate the stability of the explanations with respect to input shifts, and the fidelity with respect to increased model performance. We quantitatively evaluate the correlation between available pixel-wise annotations and explainability heatmaps. We show that the explanations on important tiles of the whole slide correlate with tissue changes between healthy regions and lesions, but do not exactly behave like a human annotator. This result is coherent with the model training strategy.
3D-Morphomics, Morphological Features on CT scans for lung nodule malignancy diagnosis
Jul 27, 2022
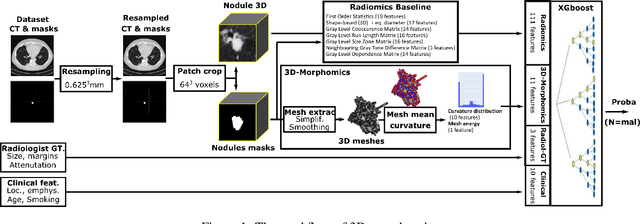
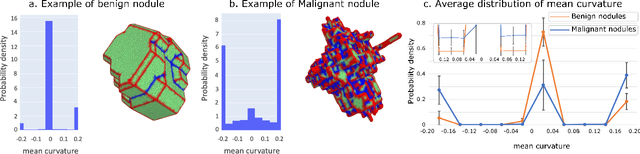
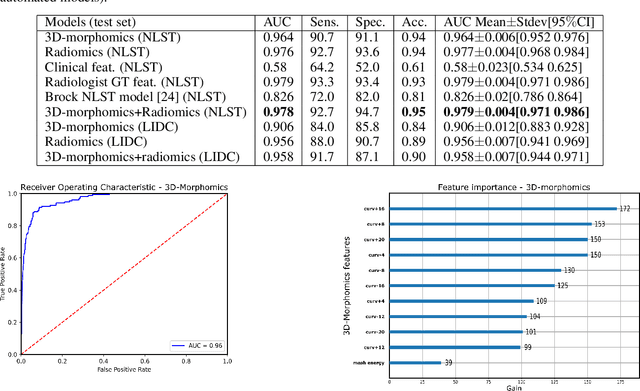
Abstract:Pathologies systematically induce morphological changes, thus providing a major but yet insufficiently quantified source of observables for diagnosis. The study develops a predictive model of the pathological states based on morphological features (3D-morphomics) on Computed Tomography (CT) volumes. A complete workflow for mesh extraction and simplification of an organ's surface is developed, and coupled with an automatic extraction of morphological features given by the distribution of mean curvature and mesh energy. An XGBoost supervised classifier is then trained and tested on the 3D-morphomics to predict the pathological states. This framework is applied to the prediction of the malignancy of lung's nodules. On a subset of NLST database with malignancy confirmed biopsy, using 3D-morphomics only, the classification model of lung nodules into malignant vs. benign achieves 0.964 of AUC. Three other sets of classical features are trained and tested, (1) clinical relevant features gives an AUC of 0.58, (2) 111 radiomics gives an AUC of 0.976, (3) radiologist ground truth (GT) containing the nodule size, attenuation and spiculation qualitative annotations gives an AUC of 0.979. We also test the Brock model and obtain an AUC of 0.826. Combining 3D-morphomics and radiomics features achieves state-of-the-art results with an AUC of 0.978 where the 3D-morphomics have some of the highest predictive powers. As a validation on a public independent cohort, models are applied to the LIDC dataset, the 3D-morphomics achieves an AUC of 0.906 and the 3D-morphomics+radiomics achieves an AUC of 0.958, which ranks second in the challenge among deep models. It establishes the curvature distributions as efficient features for predicting lung nodule malignancy and a new method that can be applied directly to arbitrary computer aided diagnosis task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge