Floriane Montanari
From slides (through tiles) to pixels: an explainability framework for weakly supervised models in pre-clinical pathology
Feb 03, 2023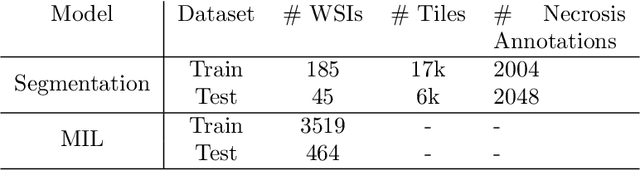
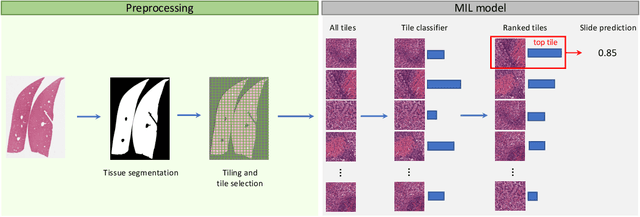
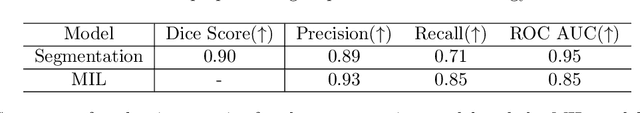
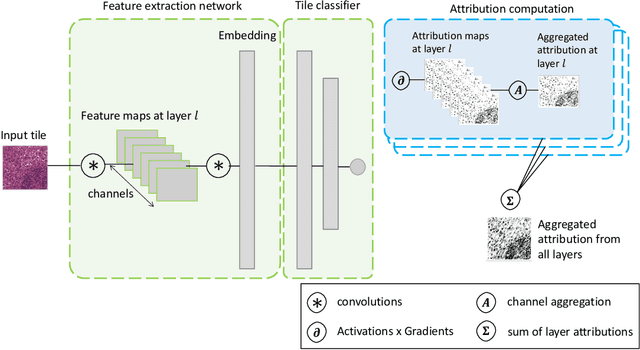
Abstract:In pre-clinical pathology, there is a paradox between the abundance of raw data (whole slide images from many organs of many individual animals) and the lack of pixel-level slide annotations done by pathologists. Due to time constraints and requirements from regulatory authorities, diagnoses are instead stored as slide labels. Weakly supervised training is designed to take advantage of those data, and the trained models can be used by pathologists to rank slides by their probability of containing a given lesion of interest. In this work, we propose a novel contextualized eXplainable AI (XAI) framework and its application to deep learning models trained on Whole Slide Images (WSIs) in Digital Pathology. Specifically, we apply our methods to a multi-instance-learning (MIL) model, which is trained solely on slide-level labels, without the need for pixel-level annotations. We validate quantitatively our methods by quantifying the agreements of our explanations' heatmaps with pathologists' annotations, as well as with predictions from a segmentation model trained on such annotations. We demonstrate the stability of the explanations with respect to input shifts, and the fidelity with respect to increased model performance. We quantitatively evaluate the correlation between available pixel-wise annotations and explainability heatmaps. We show that the explanations on important tiles of the whole slide correlate with tissue changes between healthy regions and lesions, but do not exactly behave like a human annotator. This result is coherent with the model training strategy.
Explaining, Evaluating and Enhancing Neural Networks' Learned Representations
Feb 18, 2022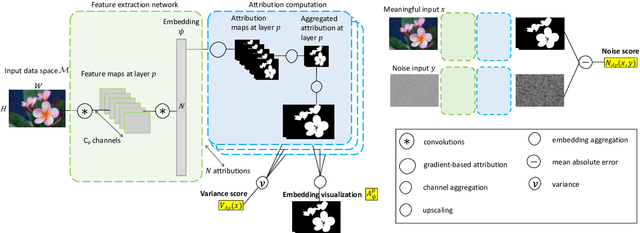
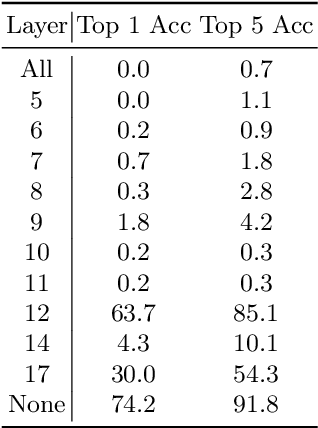
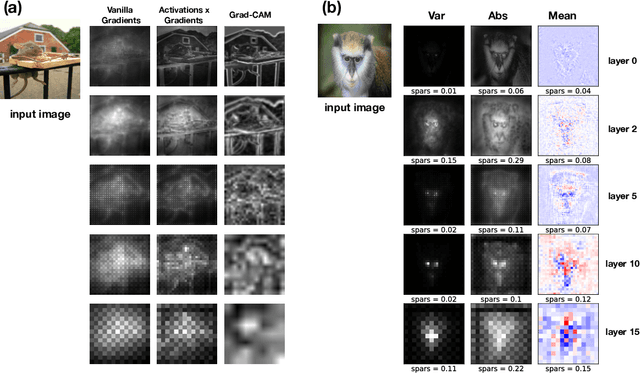
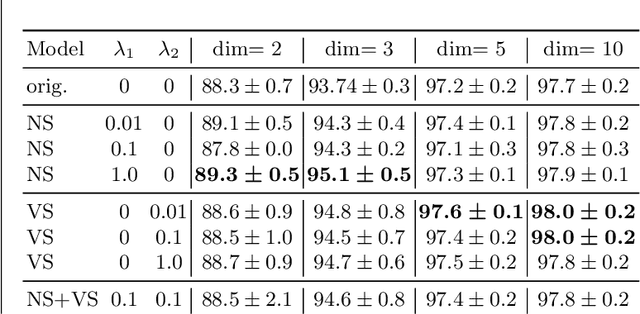
Abstract:Most efforts in interpretability in deep learning have focused on (1) extracting explanations of a specific downstream task in relation to the input features and (2) imposing constraints on the model, often at the expense of predictive performance. New advances in (unsupervised) representation learning and transfer learning, however, raise the need for an explanatory framework for networks that are trained without a specific downstream task. We address these challenges by showing how explainability can be an aid, rather than an obstacle, towards better and more efficient representations. Specifically, we propose a natural aggregation method generalizing attribution maps between any two (convolutional) layers of a neural network. Additionally, we employ such attributions to define two novel scores for evaluating the informativeness and the disentanglement of latent embeddings. Extensive experiments show that the proposed scores do correlate with the desired properties. We also confirm and extend previously known results concerning the independence of some common saliency strategies from the model parameters. Finally, we show that adopting our proposed scores as constraints during the training of a representation learning task improves the downstream performance of the model.
Improving Molecular Graph Neural Network Explainability with Orthonormalization and Induced Sparsity
May 31, 2021
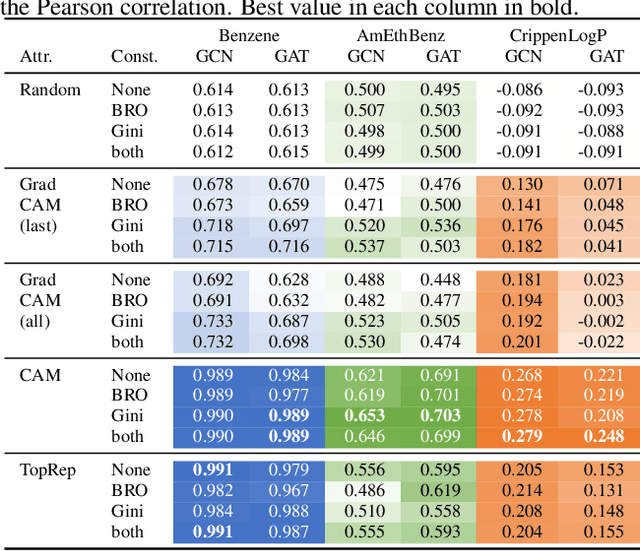
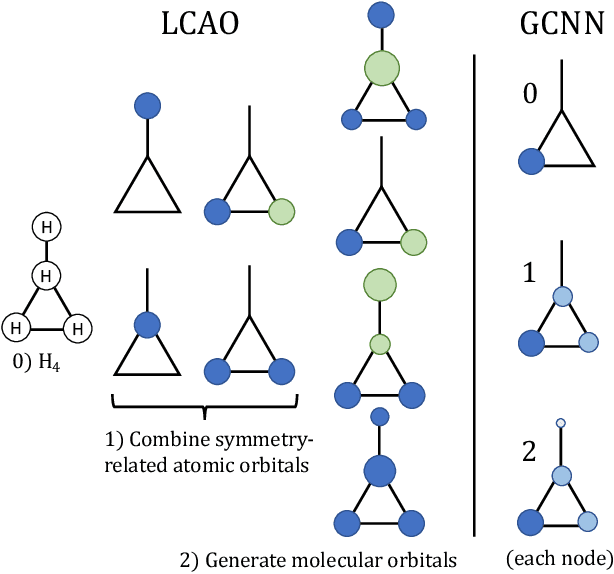
Abstract:Rationalizing which parts of a molecule drive the predictions of a molecular graph convolutional neural network (GCNN) can be difficult. To help, we propose two simple regularization techniques to apply during the training of GCNNs: Batch Representation Orthonormalization (BRO) and Gini regularization. BRO, inspired by molecular orbital theory, encourages graph convolution operations to generate orthonormal node embeddings. Gini regularization is applied to the weights of the output layer and constrains the number of dimensions the model can use to make predictions. We show that Gini and BRO regularization can improve the accuracy of state-of-the-art GCNN attribution methods on artificial benchmark datasets. In a real-world setting, we demonstrate that medicinal chemists significantly prefer explanations extracted from regularized models. While we only study these regularizers in the context of GCNNs, both can be applied to other types of neural networks
Gini in a Bottleneck: Gotta Train Me the Right Way
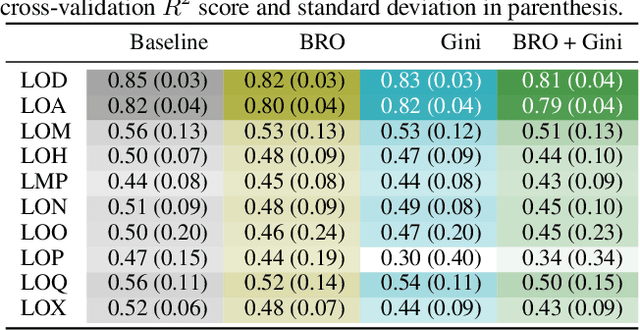
Oct 09, 2020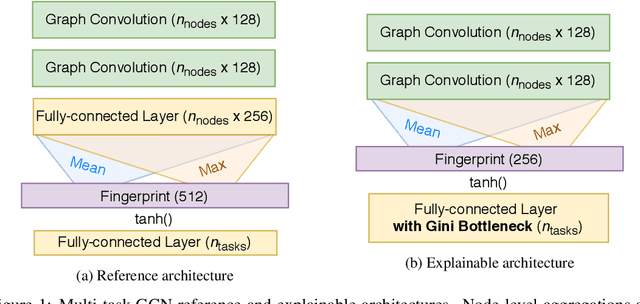
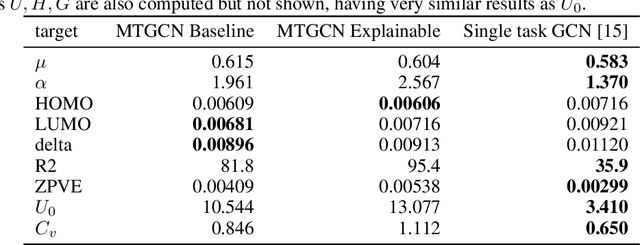
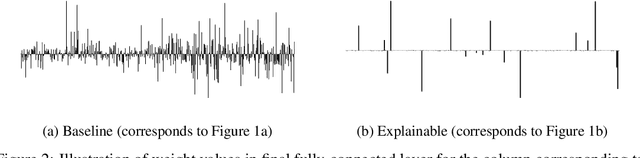
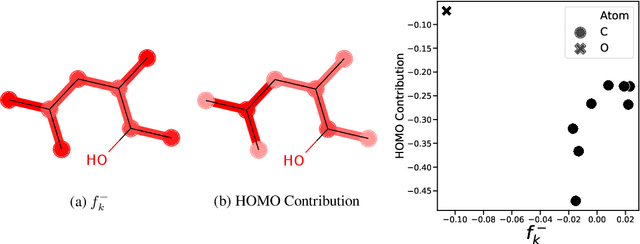
Abstract:Due to the nature of deep learning approaches, it is inherently difficult to understand which aspects of a molecular graph drive the predictions of the network. As a mitigation strategy, we constrain certain weights in a multi-task graph convolutional neural network according to the Gini index to maximize the "inequality" of the learned representations. We show that this constraint does not degrade evaluation metrics for some targets, and allows us to combine the outputs of the graph convolutional operation in a visually interpretable way. We then perform a proof-of-concept experiment on quantum chemistry targets on the public QM9 dataset, and a larger experiment on ADMET targets on proprietary drug-like molecules. Since a benchmark of explainability in the latter case is difficult, we informally surveyed medicinal chemists within our organization to check for agreement between regions of the molecule they and the model identified as relevant to the properties in question.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge