Ryan Henderson
Improving Molecular Graph Neural Network Explainability with Orthonormalization and Induced Sparsity
May 31, 2021
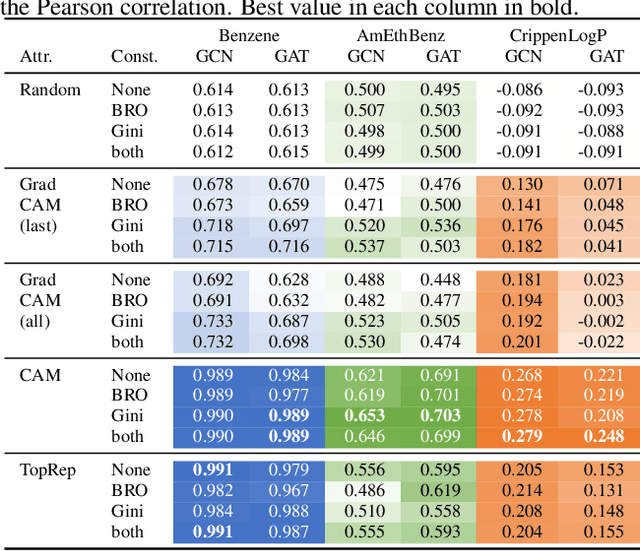
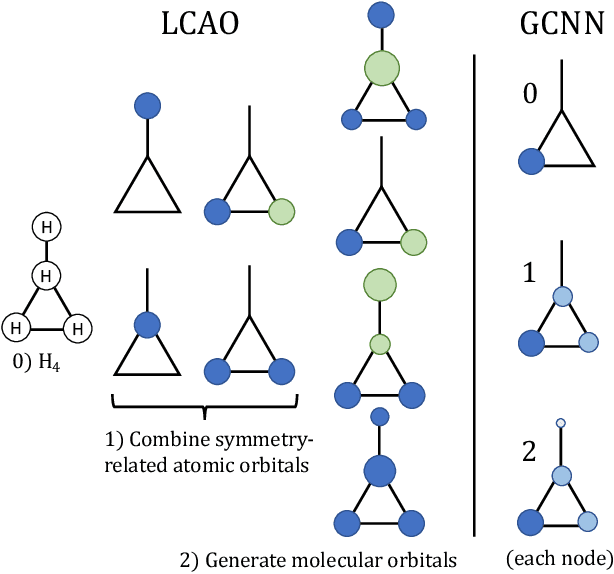
Abstract:Rationalizing which parts of a molecule drive the predictions of a molecular graph convolutional neural network (GCNN) can be difficult. To help, we propose two simple regularization techniques to apply during the training of GCNNs: Batch Representation Orthonormalization (BRO) and Gini regularization. BRO, inspired by molecular orbital theory, encourages graph convolution operations to generate orthonormal node embeddings. Gini regularization is applied to the weights of the output layer and constrains the number of dimensions the model can use to make predictions. We show that Gini and BRO regularization can improve the accuracy of state-of-the-art GCNN attribution methods on artificial benchmark datasets. In a real-world setting, we demonstrate that medicinal chemists significantly prefer explanations extracted from regularized models. While we only study these regularizers in the context of GCNNs, both can be applied to other types of neural networks
Gini in a Bottleneck: Gotta Train Me the Right Way
Oct 09, 2020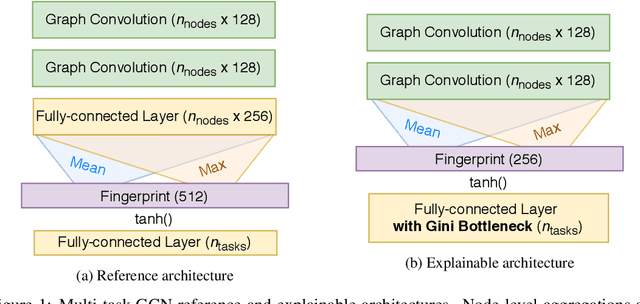
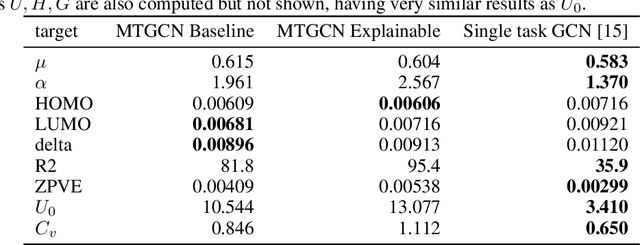
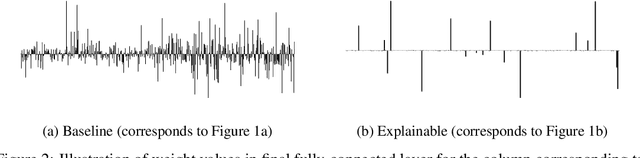
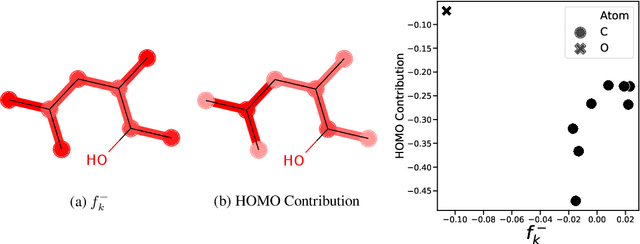
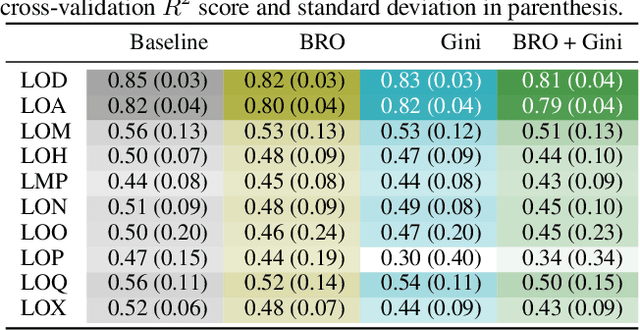
Abstract:Due to the nature of deep learning approaches, it is inherently difficult to understand which aspects of a molecular graph drive the predictions of the network. As a mitigation strategy, we constrain certain weights in a multi-task graph convolutional neural network according to the Gini index to maximize the "inequality" of the learned representations. We show that this constraint does not degrade evaluation metrics for some targets, and allows us to combine the outputs of the graph convolutional operation in a visually interpretable way. We then perform a proof-of-concept experiment on quantum chemistry targets on the public QM9 dataset, and a larger experiment on ADMET targets on proprietary drug-like molecules. Since a benchmark of explainability in the latter case is difficult, we informally surveyed medicinal chemists within our organization to check for agreement between regions of the molecule they and the model identified as relevant to the properties in question.
Picasso: A Modular Framework for Visualizing the Learning Process of Neural Network Image Classifiers
Sep 11, 2017


Abstract:Picasso is a free open-source (Eclipse Public License) web application written in Python for rendering standard visualizations useful for analyzing convolutional neural networks. Picasso ships with occlusion maps and saliency maps, two visualizations which help reveal issues that evaluation metrics like loss and accuracy might hide: for example, learning a proxy classification task. Picasso works with the Tensorflow deep learning framework, and Keras (when the model can be loaded into the Tensorflow backend). Picasso can be used with minimal configuration by deep learning researchers and engineers alike across various neural network architectures. Adding new visualizations is simple: the user can specify their visualization code and HTML template separately from the application code.
* 9 pages, submission to the Journal of Open Research Software, github.com/merantix/picasso
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge