Jean-Christophe Brisset
3D-Morphomics, Morphological Features on CT scans for lung nodule malignancy diagnosis
Jul 27, 2022
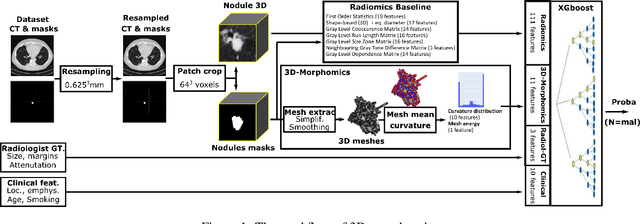
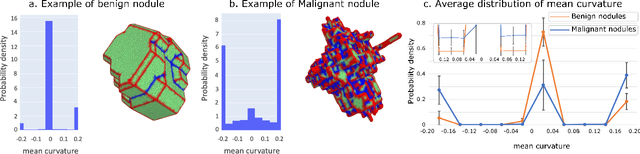
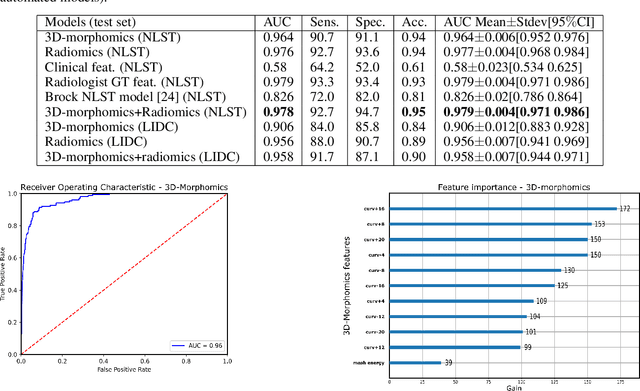
Abstract:Pathologies systematically induce morphological changes, thus providing a major but yet insufficiently quantified source of observables for diagnosis. The study develops a predictive model of the pathological states based on morphological features (3D-morphomics) on Computed Tomography (CT) volumes. A complete workflow for mesh extraction and simplification of an organ's surface is developed, and coupled with an automatic extraction of morphological features given by the distribution of mean curvature and mesh energy. An XGBoost supervised classifier is then trained and tested on the 3D-morphomics to predict the pathological states. This framework is applied to the prediction of the malignancy of lung's nodules. On a subset of NLST database with malignancy confirmed biopsy, using 3D-morphomics only, the classification model of lung nodules into malignant vs. benign achieves 0.964 of AUC. Three other sets of classical features are trained and tested, (1) clinical relevant features gives an AUC of 0.58, (2) 111 radiomics gives an AUC of 0.976, (3) radiologist ground truth (GT) containing the nodule size, attenuation and spiculation qualitative annotations gives an AUC of 0.979. We also test the Brock model and obtain an AUC of 0.826. Combining 3D-morphomics and radiomics features achieves state-of-the-art results with an AUC of 0.978 where the 3D-morphomics have some of the highest predictive powers. As a validation on a public independent cohort, models are applied to the LIDC dataset, the 3D-morphomics achieves an AUC of 0.906 and the 3D-morphomics+radiomics achieves an AUC of 0.958, which ranks second in the challenge among deep models. It establishes the curvature distributions as efficient features for predicting lung nodule malignancy and a new method that can be applied directly to arbitrary computer aided diagnosis task.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018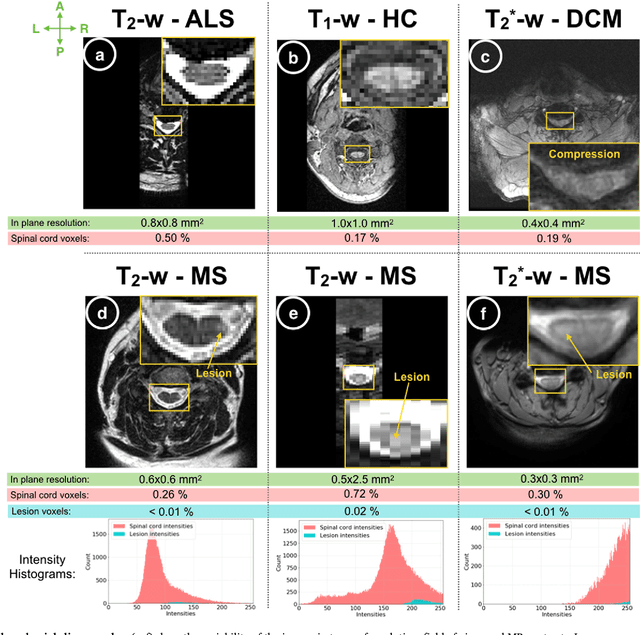
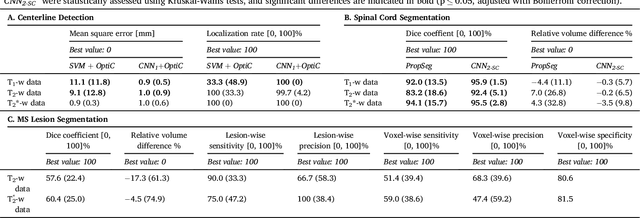
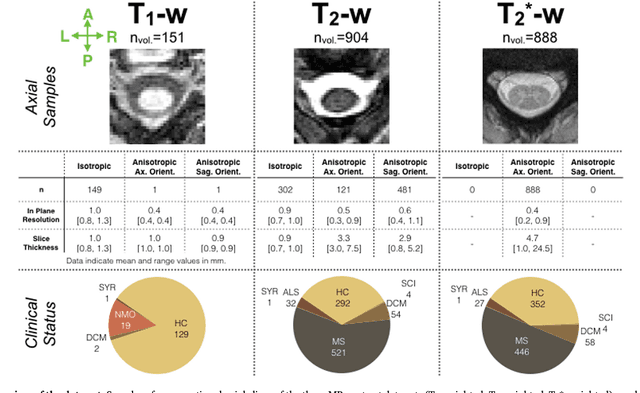
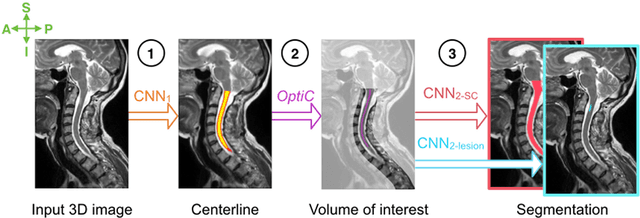
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge